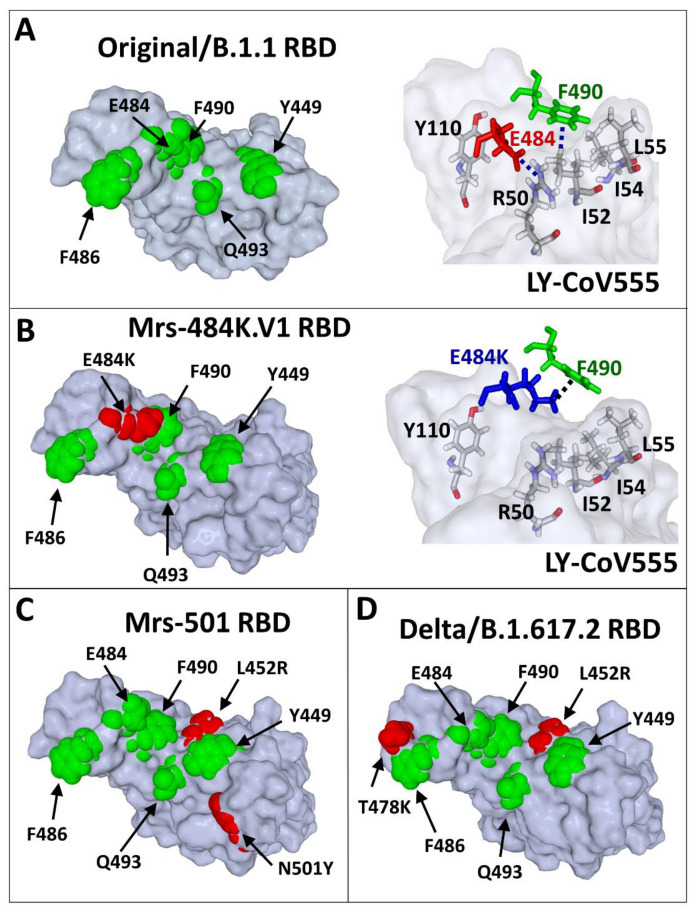
Figure 4.
Scheme showing the molecular mechanisms of nAb escape in the RBD of SARS-CoV-2 variants. (A) The epitope recognised by the LY-CoV555 nAb (pdb file #7KMG) consists of several amino acid (coloured in green) residues distributed on the surface of the RBD. The anionic carboxylic group of E484 interacts with the cationic charge of R50 (heavy chain of LY-CoV555 nAb) through an electrostatic bridge. The aromatic ring of Y490 interacts with a methyl group of I52 (heavy chain of LY-CoV555 nAb) by a CH–π interaction, which is reinforced by vicinal apolar amino acid residues (I54 and I55). (B) In the Marseille-484K.V1/R.1 variant, E484 (in red in the left panel) is mutated in E484K (in blue in the right panel). The consequence of this mutation is a shift of the side chain of E484K whose cationic group (which replaces the negative charge of E484) now forms a cation–π bond with the aromatic ring of F490. In this new context, neither E484K nor F490 can still interact with the LY-CoV555 nAb. Indeed, R50, I52, I54, and L55 of the heavy chain of the antibody are no longer involved in RBD recognition. (C) Mutational pattern of the Marseille-501/A.27 variant (L452R/N501Y). (D) Mutational pattern of the India_1 variant (L452R/T478K). The same molecular modelling method was applied to all variants (B–D) after introducing the mutations in the reference Original/B.1.1–nAb complex (PDB file #7KMG), followed by energy minimisation of the RBD and simulations of the binding reaction.