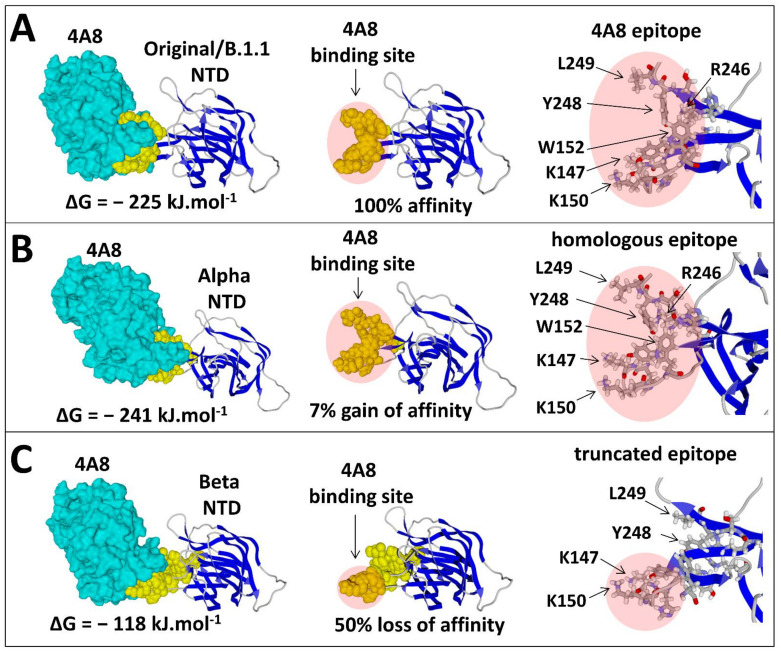
Figure 7.
Figure showing the variability of the main neutralising epitope in the NTD among virus strains. (A) Molecular mechanism of NTD recognition (Original/B.1.1 strain) by the 4A8 nAb. The NTD–nAb complex (pdb file #7C2 L) has a global affinity of −225 kJ/mol−1. The antibody clamps two distal zones of the NTD (the N3 loop with amino acid residues K147, K150, and W152) and the N5 loop (R246, Y248, and L249), which together form the main neutralising epitope of the NTD. (B) The NTD of the Alpha variant retains this crescent-shaped structure, which displays a slightly higher affinity (+ 7% compared with the Original/B.1.1 strain) for the 4A8 nAb due to the repositioning of the amino acids of the N3 loop. (C) In the case of the Betavariant, only the N3 loop part of the epitope is conserved, and therefore that the affinity for the 4A8 nAb is decreased by 50%. Such a truncated epitope may elicit a poor antibody response, consistent with seroneutralisation data.