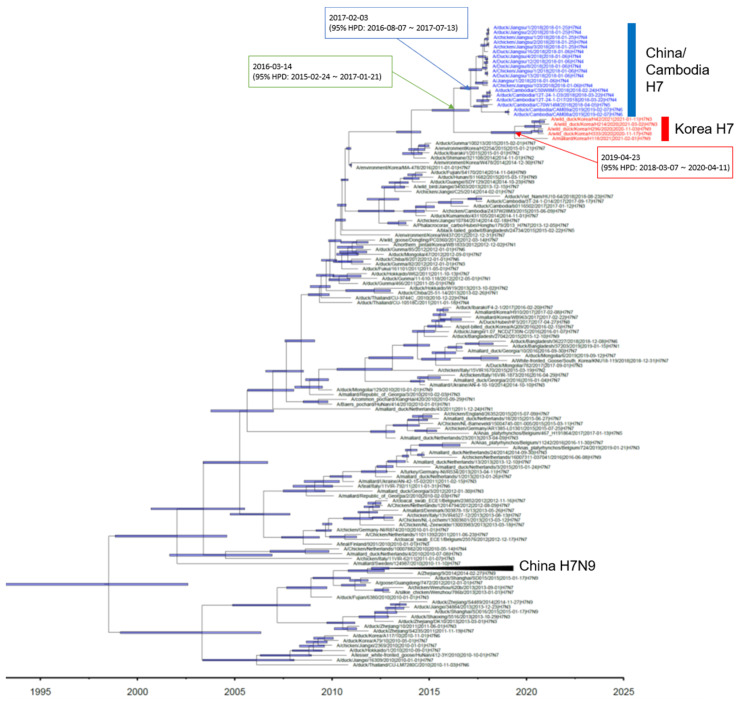
Figure 1.
Maximum clade credibility phylogeny of the HA gene sequences of H7Nx avian influenza viruses in Eurasia. We estimated the maximum clade credibility tree using a time-measured Bayesian phylogenetic model implemented in BEAST v1.10.4. The Hasegawa–Kishino–Yano nucleotide substitution model, with a discrete gamma distribution with four categories, was applied. Branch lengths represent time. Horizontal bars indicate 95% Bayesian credible intervals for estimates of common ancestry. The H7Nx viruses isolated in China and Cambodia are colored with blue. The H7Nx viruses isolated in this study are colored with red. The estimated time of the most recent common ancestry with 95% highest posterior density (HPD) is displayed on the node of cluster.