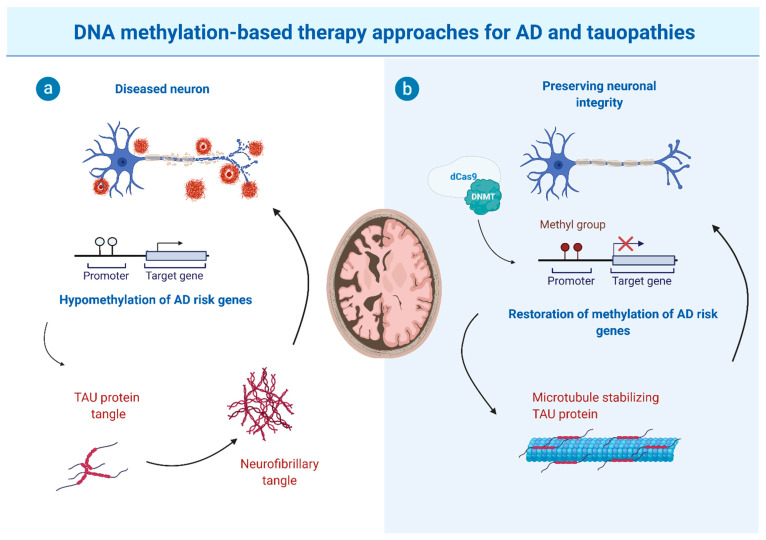
Figure 1.
Putative potential of CRISPR/dCas9 editing-based therapeutic approaches for tauopathies that display impaired methylation patterns of selected genes/key regulator elements. (a) In disease paradigms, impaired DNA methylation (e.g., hypomethylation of risk genes associated with Alzheimer’s Disease (AD) and related/other tauopathies) results in increased TAU expression, decreased TAU clearance, or mislocalization, all of which lead to the accumulation of TAU and eventually to the formation of TAU protein aggregates. Neurons affected by this TAU pathology become dysfunctional and decay, eventually leading to impaired cognitive function and neurodegeneration. (b) CRISPR/dCas9 editing approaches may restore methylation patterns of AD and tauopathy risk genes, preventing abnormal production or modification of TAU protein and NFT formation, preserving the physiological function of TAU (i.a. microtubule stabilization) and preventing or partially reverse brain damage and disease progression. Possible genetic and non-genetic interventions could be (i) drug-induced modulation of methylation patterns, (ii) gene-replacement or RNAi-based gene therapy, or (iii) site/gene-specific modulation of methylation, e.g., as depicted, site-specific methylation via dCas9-directed DNMT targeting. This figure produced by using BioRender.com with a respective publication licence, provided by the Biology department of the RWTH Aachen University.