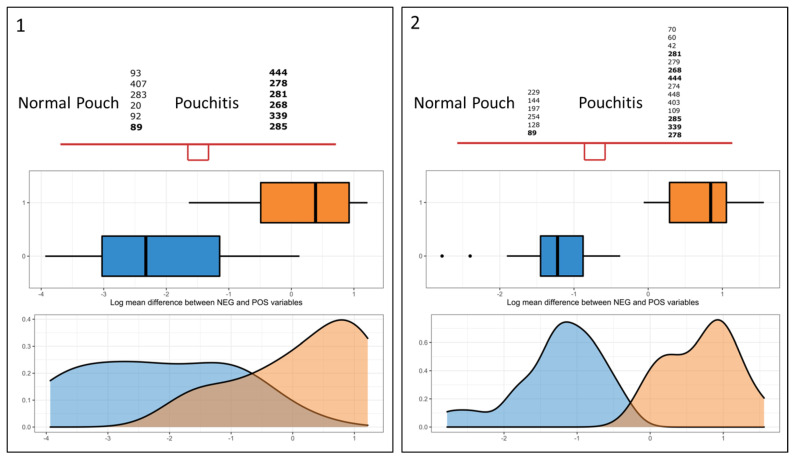
Figure 5.
Differential microbial abundance between the normal pouch and pouchitis groups by using two compositional data analysis (CoDA) approaches, namely coda-lasso (panel 1) and clr-lasso (panel 2). The “selbal-like” plots show the feature identifiers for the clr-lasso- and coda-lasso-selected taxa that have greater abundance in “class 1” samples, i.e., the pouchitis samples (id in “A” list) and those more abundant in “class 0” samples (normal pouch, identifiers in “B” list). Bold numbers in the two top panels represent common features between methods; identifiers are ordered according to their absolute coefficients (lowest to highest and in top-down direction). Middle and bottom panels show, respectively, boxplots of the scores (or balances) as calculated for each sample within each sample group and density plots of such balances. “Balance” resumes the mean log-transformed abundances of the taxa in the comparing groups.