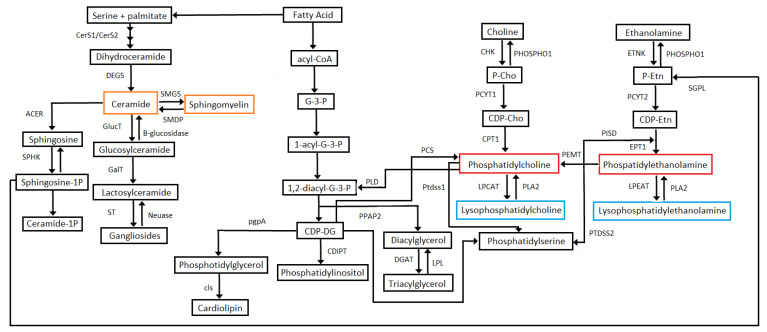
Figure 7.
Schematic Representation of lipid metabolic pathways. Metabolite abbreviations: G-3-P, sn-Glycerol 3-Phosphate; 1-acyl-G-3-P, 1-Acyl-sn-glycerol 3-phosphate; 1,2-diacyl-G-3-P, 1,2-Diacyl-sn-glycerol 3-phosphate; CDP-DG, CDP-1,2-diacylglycerol; P-Cho, Choline Phosphate; CDP-Cho, CDP-choline; P-Etn, Phosphoethanolamine; CDP-Etn, CDP-ethanolamine. Enzyme abbreviations: CerS1/CerS2, Ceramide Synthase 1/Ceramide Synthase 2; DEGS, sphingolipid delta-4 desaturase; SMGS, sphingomyelin synthase; SMDP, Sphingomyelin Phosphodiesterase; ACER, alkaline ceramidase; SPHK, sphingosine kinase; GlucT, Glucosylceramide-Transferase; GalT, Galactosylceramide-Transferase; ST, Sialyl-Transferase; Neusase, Neuraminase; pgpA, phosphatidylglycerophosphatase A; cls, cardiolipin synthase; PLD, Phospholipase D1/2; PPAP2, phosphatidate phosphatase; CDIPT, CDP-diacylglycerol-inositol 3-phosphatidyltransferase; CHK, choline kinase; PHOSPHO1, phosphoethanolamine/phosphocholine 586 phosphatase; PCYT1, choline-phosphate cytidylyltransferase; CPT1, diacylglycerol cholinephosphotransferase; PCS, phosphatidylcholine synthase; DGAT, diacylglycerol O-acyltransferase; LPC, lipoprotein lipase; Ptdss1, phosphatidylserine synthase 1; LPCAT, lysophosphatidylcholine 650 acyltransferase; PLA2, secretory phospholipase A2; ETNK, Ethanolamine Kinase; PCYT2, ethanolamine-phosphate cytidylyltransferase; PISD, phosphatidylserine decarboxylase; EPT1, ethanolaminephosphotransferase; PEMT, phosphatidylethanolamine N-methyltransferase; LPEAT, lysophospholipid acyltransferase; PTDSS2, phosphatidylserine synthase 2. Lipid species analysed in the present study: Orange rectangles = sphingolipids, red rectangles = phospholipids, blue rectangles = lysophospholipids.