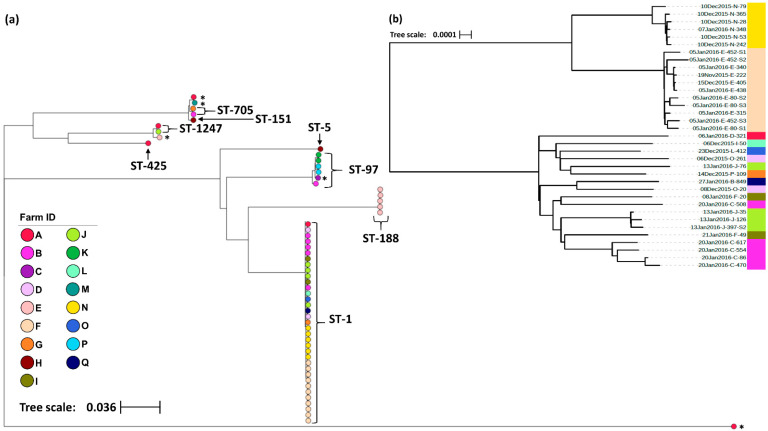
Figure 2.
(a) Maximum-likelihood phylogeny generated from core single nucleotide polymorphisms across the 57 S. aureus isolates. All isolates were collected from bovine raw milk samples in New Zealand and have been labelled with their sequence type (ST) as identified by seven MLST genes. Those isolates with an asterisk (*) indicate isolates whose ST remained undetermined. (b) Maximum-likelihood phylogeny generated from core single nucleotide polymorphisms across the 35 S. aureus ST-1 isolates identified in (a). Isolate IDs identify the date the sample was collected (dd/mm/yyyy), the farm from which it was collected from (A–Q), and the animal ID number. In both trees, the color of the terminal tree nodes also corresponds to the farm from which the isolate was collected. Both figures were created using the online tool Microreact [37].