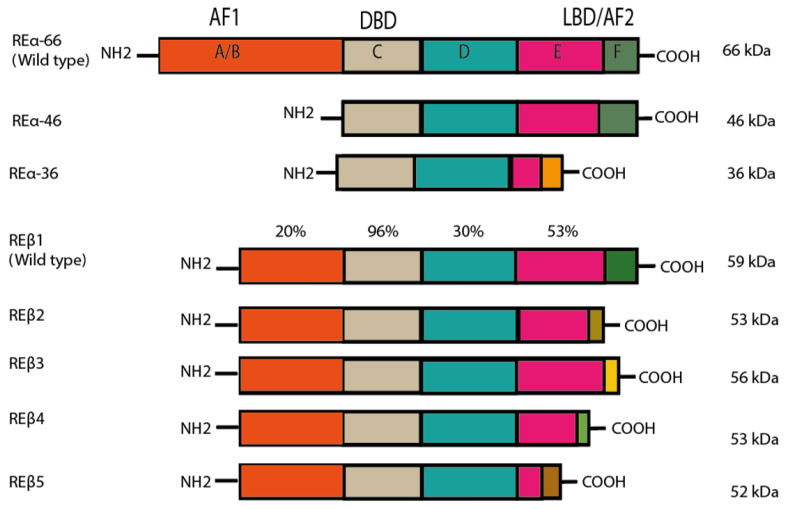
Figure 1.
Structure of estrogen receptors (ERs), as modified by Jia et al. (2015). ER⍺ and ERβ have the same structural regions (A–F), although the degree of similarity of the domains varies. The A/B amino-terminal region contains the amino-terminal domain and the ligand-independent AF-1 domain. These domains are responsible for the recruitment of coregulators (coactivators and corepressors). The C region is the binding domain of DNA (DBD), while D is known as the hinge region. The latter also encompasses part of the ligand-dependent activation function (AF) domain and the nuclear localization signal. The C-terminus region, comprising E and F, contains the ligand binding domain (LBD) and ligand-dependent AF-2. The main isoforms of ERα are portrayed, with their respective molecular weight. A difference can be appreciated in region F of ERα36, which is caused by the transcription of exon 9. In the case of ERβ, each isoform presents variations in the F domain due to the shuffling of exon 8. Reprinted from ref. [5]. Best Pract Res Clin Endocrinol Metab, 29(4), Jia M.; Dahlman-Wright K.; Gustafsson JÅ. Estrogen receptor alpha and beta in health and disease, pp. 557–568. Copyright 2015, with permission from Elsevier.