Figure 4.
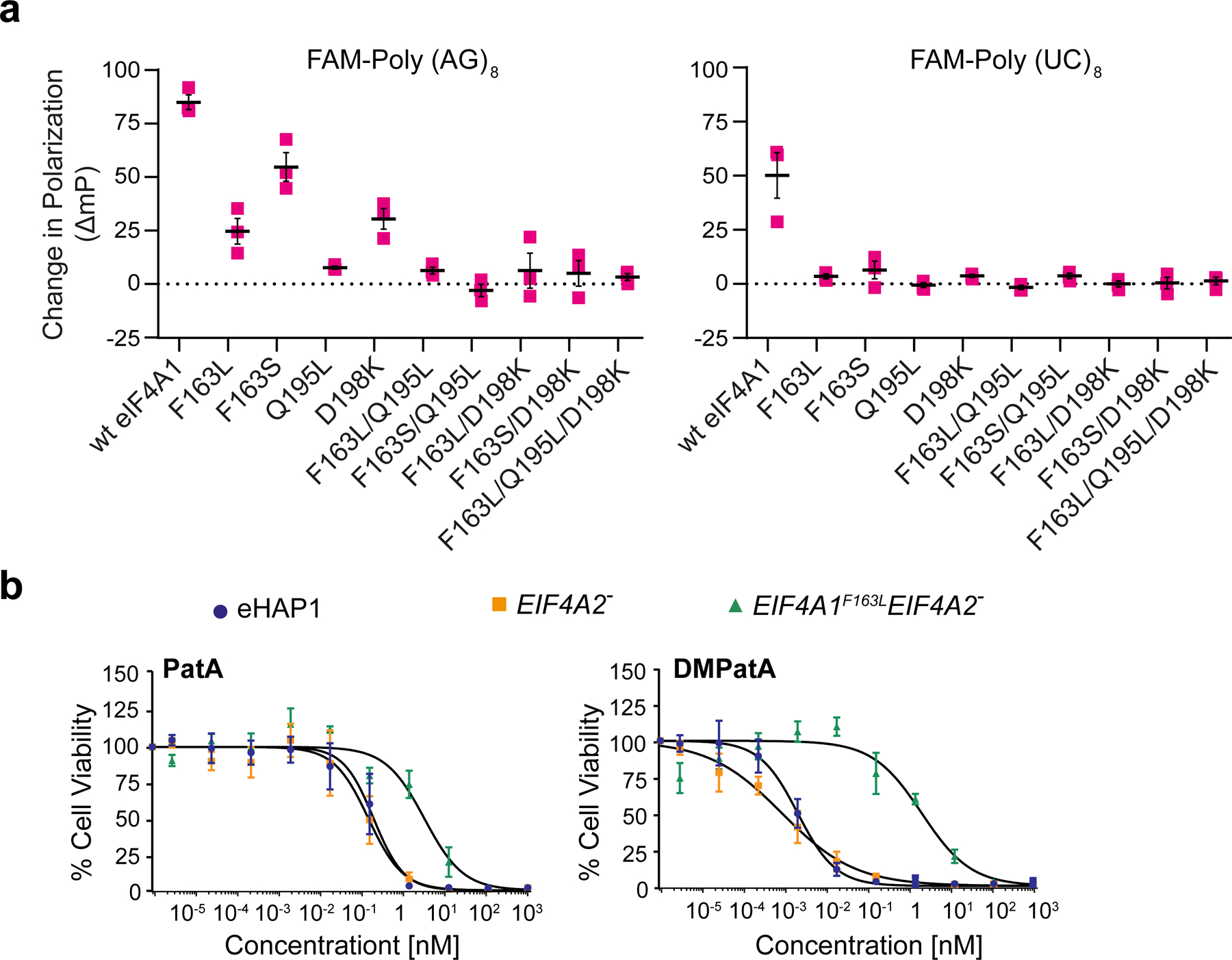
Assessing eIF4A1 amino acid requirement for DMPatA binding. a. Consequence of amino acids substitutions in eIF4A1 on DMPatA-stimulated RNA binding. The ΔmP (relative to DMSO controls) in the presence of 2.5 μM DMPatA using FAM-poly (AG)8 or FAM-poly (UC)8 RNA with the indicated eIF4A1 mutants is presented. n=3 ± SEM. b. Cytotoxicity of PatA and analogs towards wt (blue circles), EIF4A2− (orange squares), and EIF4A1F163L EIF4A2− (green triangles) eHAP1 cells. Cell were exposed to the indicated concentrations of compound for 2 days and viability was measured using the SRB assay. The IC50’s of compounds towards the test cell lines were: PatA/eHAP, 0.2 ± 0.05 nM; PatA/EIF4A2−, 0.1 ± 0.05 nM; PatA/EIF4A1F163LEIF4A2−, 3.2 ± 0.9 nM; DMPatA/eHAP, 1.9 ± 0.4 pM; DMPatA/EIF4A2−, 0.79 ± 0.23 pM; DMPatA/EIF4A1F163LEIF4A2−, 2± 0.69 nM, n = 4 ± SEM. See also Figures S4 and S5.
