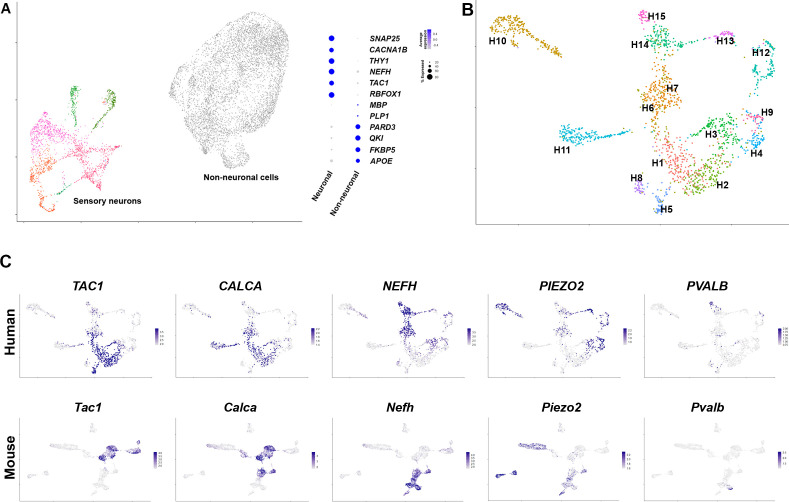
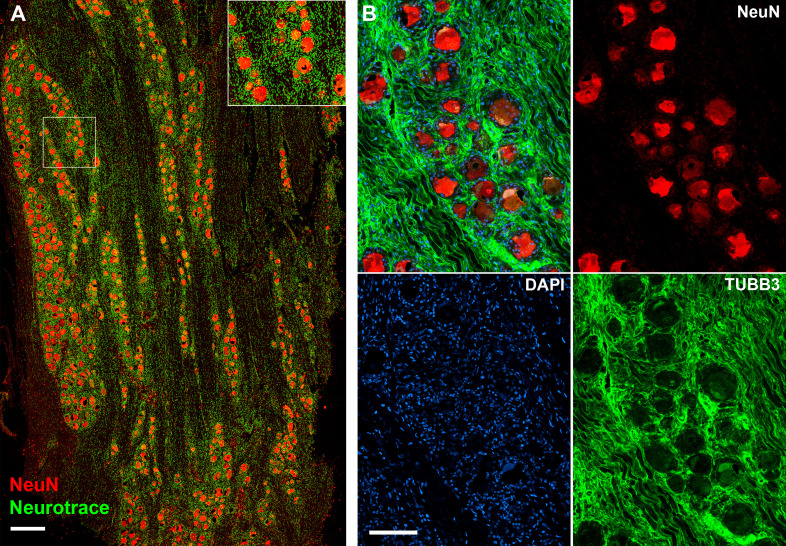
Figure 1. Diverse classes of human dorsal root ganglia (DRG) neurons revealed by single nuclear transcriptomics.
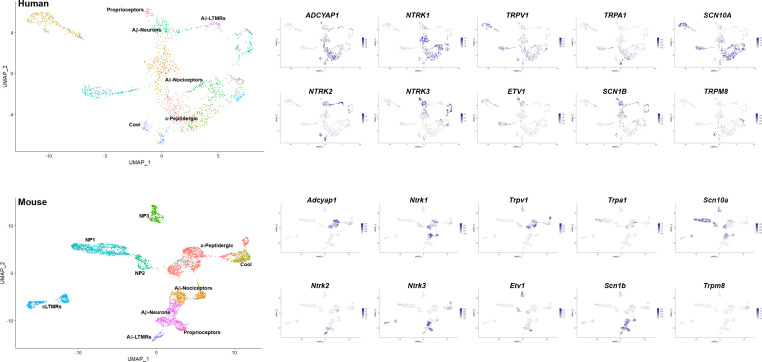
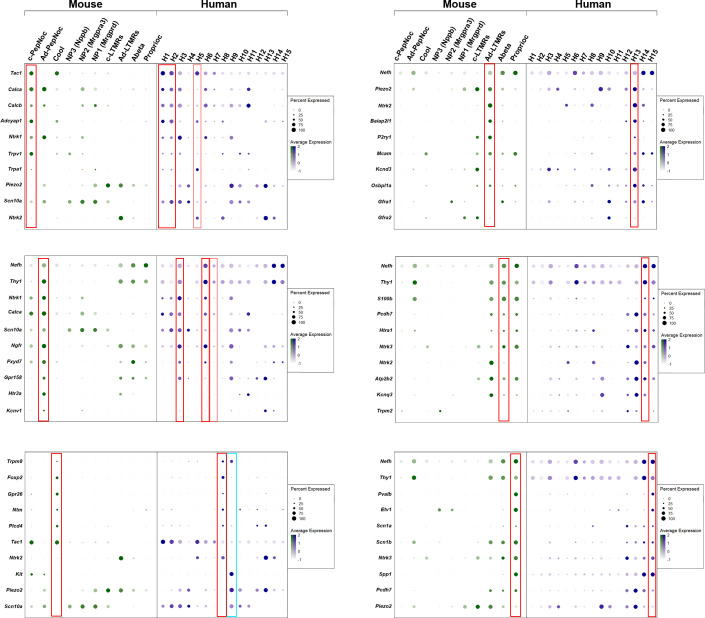
(A) Universal manifold (UMAP) representation of graph-based co-clustered snRNA sequences from human DRG nuclei reveal two well separated groups corresponding to sensory neurons (colored) and non-neuronal cells (gray). To the right, a dotplot highlights the expression of markers that help distinguish these groups of cells (see also Figure 1—figure supplement 2A for more information about preliminary analysis). (B) Reanalyzing 1837 neuronal nuclei clusters human DRG neurons into transcriptomically distinct groups that have been differentially colored. (C) Similarity in expression of differentially expressed genes between human and mouse neuronal types may help functional classification of neuronal types: UMAP representation of human DRG neurons showing relative expression level (blue) of diagnostic markers. For comparison UMAP representation of mouse neurons (Renthal et al., 2020) showing the relative expression patterns of the same markers. In combination, the expression patterns of these and other genes (Figure 1—figure supplements 2 and 4, Figure 1—figure supplement 5) were used to tentatively match several human and mouse transcriptomic classes (Figure 1—figure supplement 4).