Figure 2. GOF PIK3CA mutations and LOF CCM gene mutations co-exist in the same cell in human CCMs.
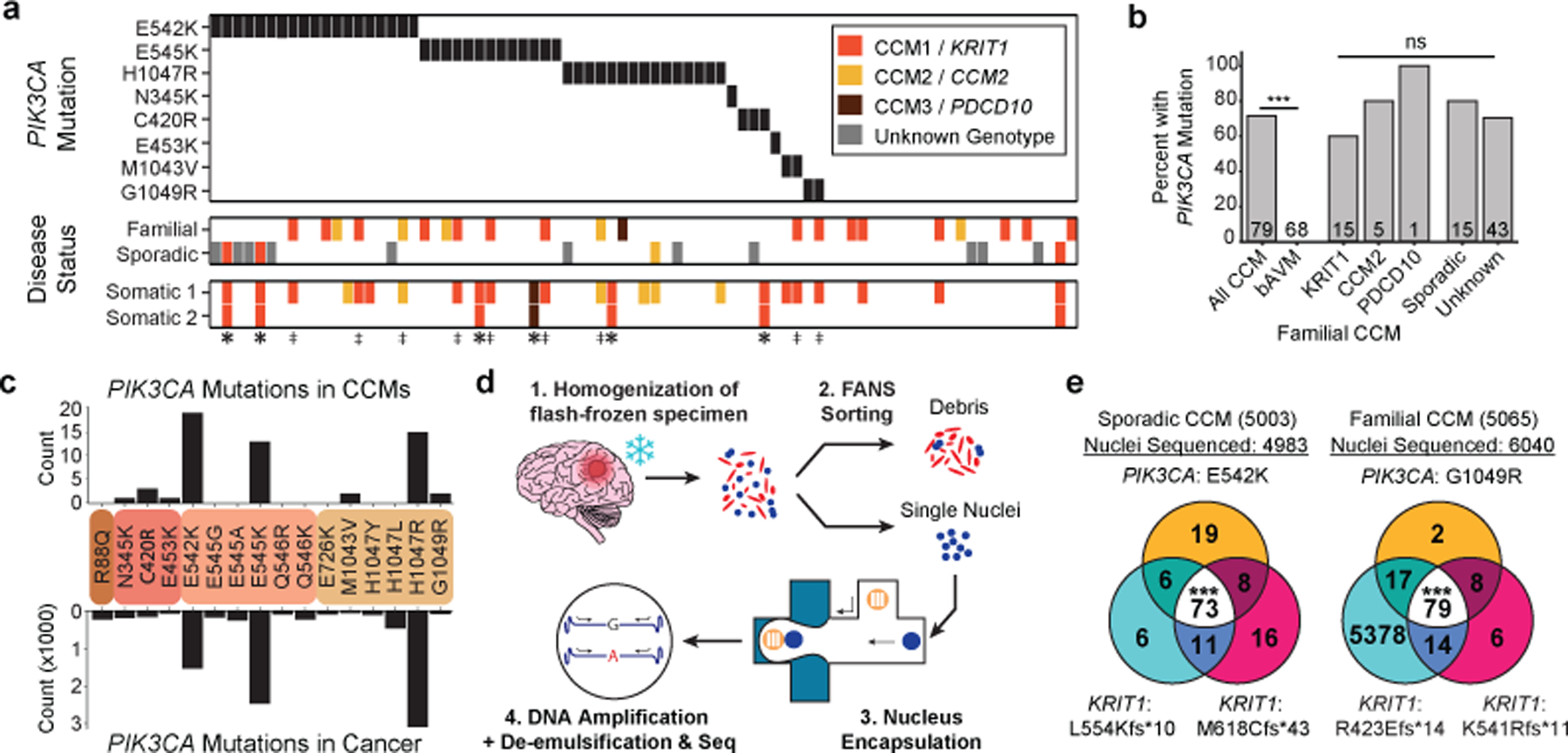
a, A schematic summary of the somatic PIK3CA mutations, the germline mutations in KRIT1, CCM2 and PDCD10, and the somatic mutations in KRIT1, CCM2 and PDCD10 as identified in 79 human CCMs via bulk sequencing of frozen resected tissue is shown. Color denotes the affected CCM gene. Samples listed as neither familial nor sporadic are deidentified banked CCMs lacking either clinical information or genetic evidence supporting either classification. ‡ indicates familial CCMs with an activating mutation in PIK3CA and both germline and somatic mutations in a CCM gene. indicates known or presumed sporadic CCMs with an activating mutation in PIK3CA and two somatic mutations in a CCM gene. b, Percentage of lesions with an activating mutation in PIK3CA present in all sequenced CCMs vs. control brain AVMs, all three forms of familial CCMs and sporadic CCMs. The value inside the bar shows the number of samples in the corresponding group. c, The distributions of the 16 most common somatic PIK3CA mutations identified in human CCMs (top) and cancer (bottom) as reported in the COSMIC database are shown. Colored boxes represent domains in PIK3CA in order from left to right: Adaptor BD, RAS BD, C2, Kinase. d, Schematic of workflow for processing frozen surgically-resected human CCM lesions for single-nucleus DNA sequencing. e, Representative data for sporadic and familial CCMs detailing the number of nuclei with each combination of PIK3CA and CCM mutations. p was determined by two-tailed chi-squared test between the observed and expected triple mutant nuclei predicted by a Poisson distribution (see Methods). ns indicates p not significant, p>0.05; ***indicates p<1e−16.
