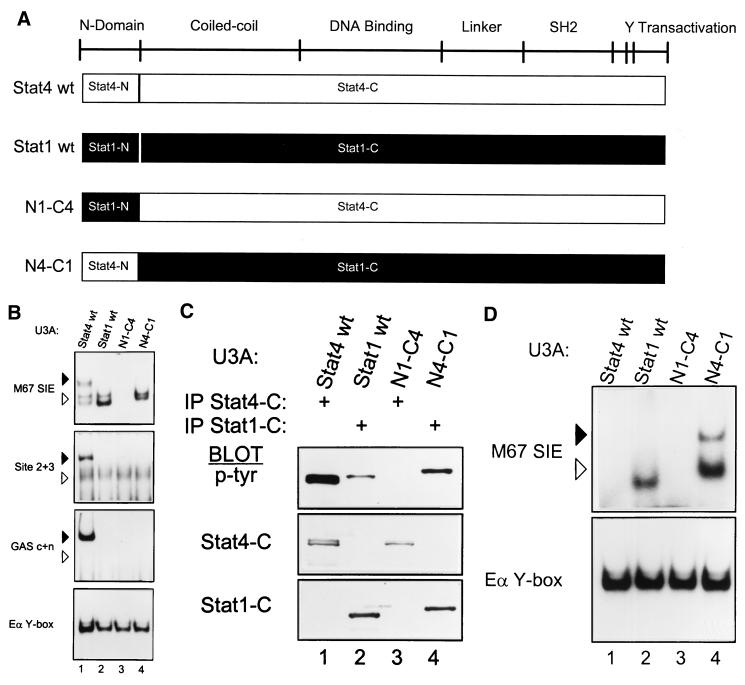
FIG. 3.
Functional noninterchangeability of Stat1 and Stat4 N domains. (A) Schematic of STAT structure showing the general positions of several domains (3). Below are diagrams of wild-type Stat4 (Stat4 wt) and Stat1 (Stat1 wt) and of two chimeras. In the N1-C4 chimera, the N domain of Stat1 (Stat1-N) is placed onto the Stat4 carboxy terminus (Stat4-C); in the N4-C1 chimera, the N domain of Stat4 (Stat4-N) is placed onto the Stat1 carboxy terminus (Stat1-C). cDNAs encoding these chimeras were placed into the retrovirus used for expression of the Stat4 mutants as described in Fig. 1. (B) U3A cells were infected with retroviruses expressing wild-type Stat4 (Stat4 wt) or Stat1 (Stat1 wt) and the N1-C4 and N4-C1 chimeras as indicated. Retrovirally infected cells were purified by cell sorting, treated with IFN-α, and nuclear extracts were prepared and analyzed by EMSA with the indicated probes as described in the legend to Fig. 1B. Arrowheads are positioned to indicate the relative migration positions between gels: solid arrowheads indicate lower mobility, and open arrowheads indicate higher mobility. Similar data were obtained in two other independent experiments. (C) The cells described for panel B were treated with IFN-α. Whole-cell extracts were prepared and immunoprecipitated with antibody to the Stat4 carboxy terminus (IP Stat4-C) or with antibody to the Stat1 carboxy terminus (IP Stat1-C) as indicated, and Western analysis was done for phosphotyrosine incorporation with the antiphosphotyrosine antibody RC20 (p-Tyr) as described above. Blots were stripped and reprobed with antimurine Stat4 antibody (Stat4-C) and antimurine Stat1 antibody (Stat1-C). (D) The cells described in the legend to panel B were treated with IFN-γ (1,000 U/ml) for 20 min. Nuclear extracts were prepared and analyzed by EMSA with the indicated probes.