Figure 1. The FA-BRCA Pathway.
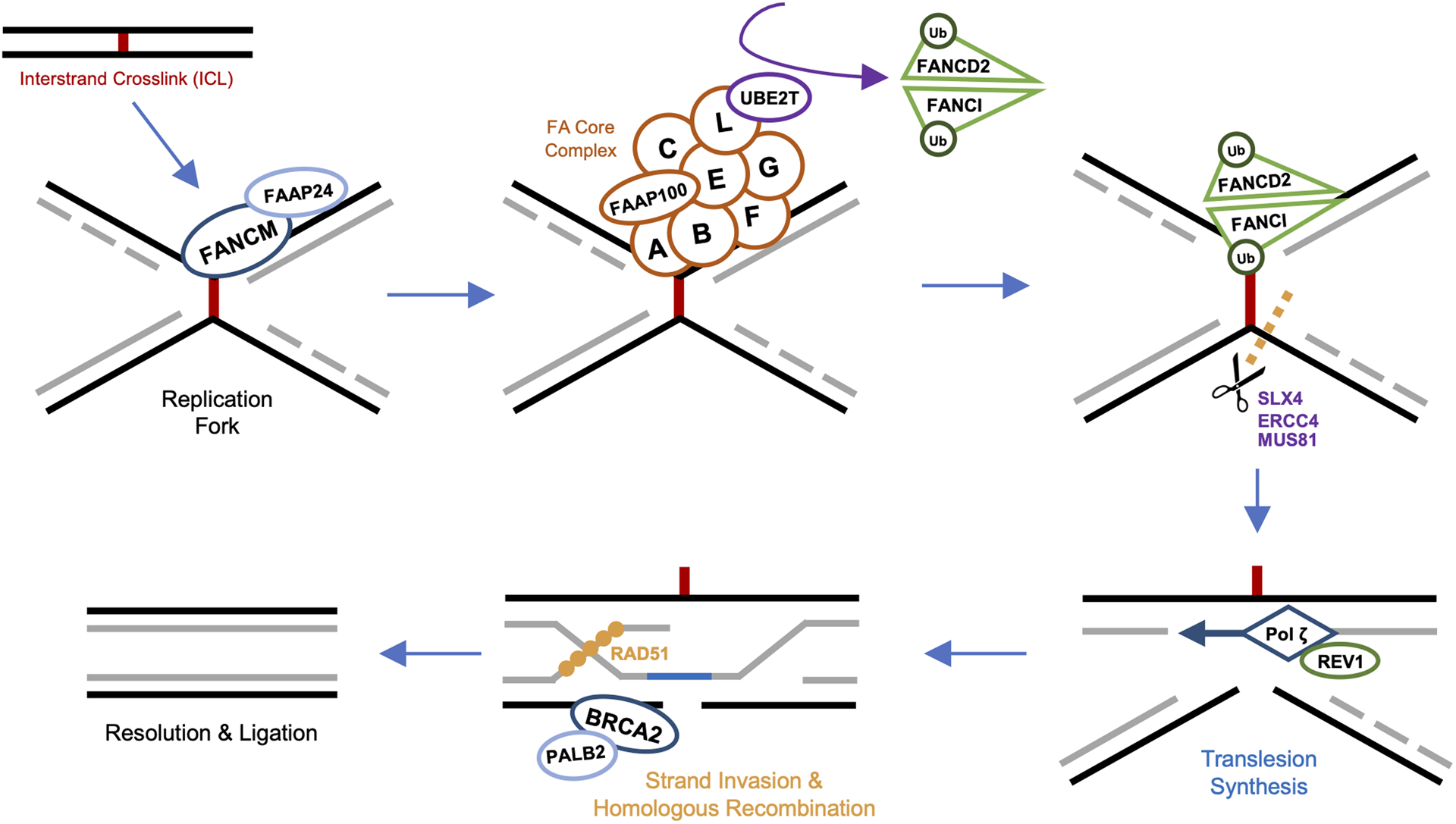
Interstrand crosslink (ICL) stalled replication forks are recognized by FANCM and its binding partner FAAP24, allowing for precise localization of the FA core complex to damaged chromatin. The FA core complex, a multimeric ubiquitin ligase, is comprised of at least 8 subunits including FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL and FAAP100. Once bound to the FANCM/FAAP24 docking platform, FANCL of the core complex associates with E2-conjugating ligase UBE2T (FANCT) to monoubiquitinate a heterodimer of FANCD2 and FANCI (FANCD2-I). Ubiquitination of FANCD2-I triggers assembly of the downstream ICL repairosome, with recruitment of SLX4 (FANCP), activating various endonucleases, including ERCC4 (FANCQ), MUS81, and SLX1. Cleavage at ICL sites by these enzymes separates the DNA strands, to which the FA core complex recruits REV1, REV3, and REV7 (FANCV) to form the translesion synthesis polymerase complex (REV1–pol ζ). Pol ζ bypasses the ICL, with the nascent strand re-establishing one of the original DNA duplexes. The double-stranded break of the remaining damaged duplex is then repaired via homologous recombination. First, BRCA2 (FANCD1) and its binding partner PALB2 (FANCN) localize to the break. BRCA2 initiates the homologous strand exchange process by recruiting recombinase RAD51 (FANCR), which facilitates invasion of the cleaved strand into the intact sister chromatid. This RAD51-guided recombination filament searches for and identifies the complementary sister sequence upon which repair synthesis and ligation are completed.
