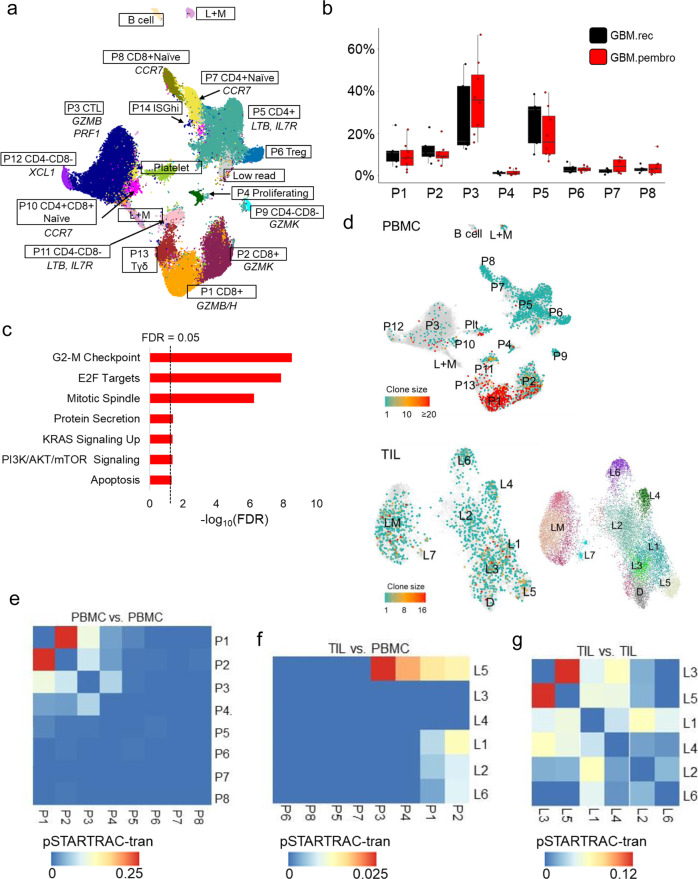
Fig. 3. The phenotype and clonal distribution of intratumoral vs. peripheral T cells in recurrent glioblastoma patients.
a A UMAP projection of the lymphoid compartment of the peripheral blood of recurrent GBM patients analyzed using scRNAseq (n = 56,444 cells from 5 GBM.rec and 8 GBM.pembro patients and two healthy donor controls). b The proportions of each lymphoid clusters in a in GBM.rec (black) and GBM.pembro (red) samples. c MSigDB Hallmark genesets showing significant overlap with the genes upregulated in the GBM.pembro PBMC (FDR values, two-sided fisher exact test). d T cell clone sizes as estimated by the TCRβ clones detected in the PBMCs (top) and TILs (bottom) using TRUST4. e, f, g Heatmaps showing the STARTRAC analysis of the shared TCRs among the PBMC clusters (e), across the PBMC and TIL clusters (f), and among the TIL clusters (g). Shared clones across two clusters indicates potential transition from one cluster to the other (non-directional). The higher the fraction of the shared TCR, the more likely that the T cells in the two clusters transition from one to the other. Source data are provided as a Source Data file or in Supp. Data 4 and 5. In all boxplots, the median is indicated by the line within the box and the 25th and 75th percentiles indicated by the lower and upper bounds of the box. The upper and lower lines above and below the boxes represent the whiskers. The gene set enrichment P values were calculated using two-sided Fisher exact test with FDR adjustment for multiple comparisons.