Figure 4. RNA and DNA modifications regulate R-loop homeostasis during gene transcription and cellular response to DNA damage.
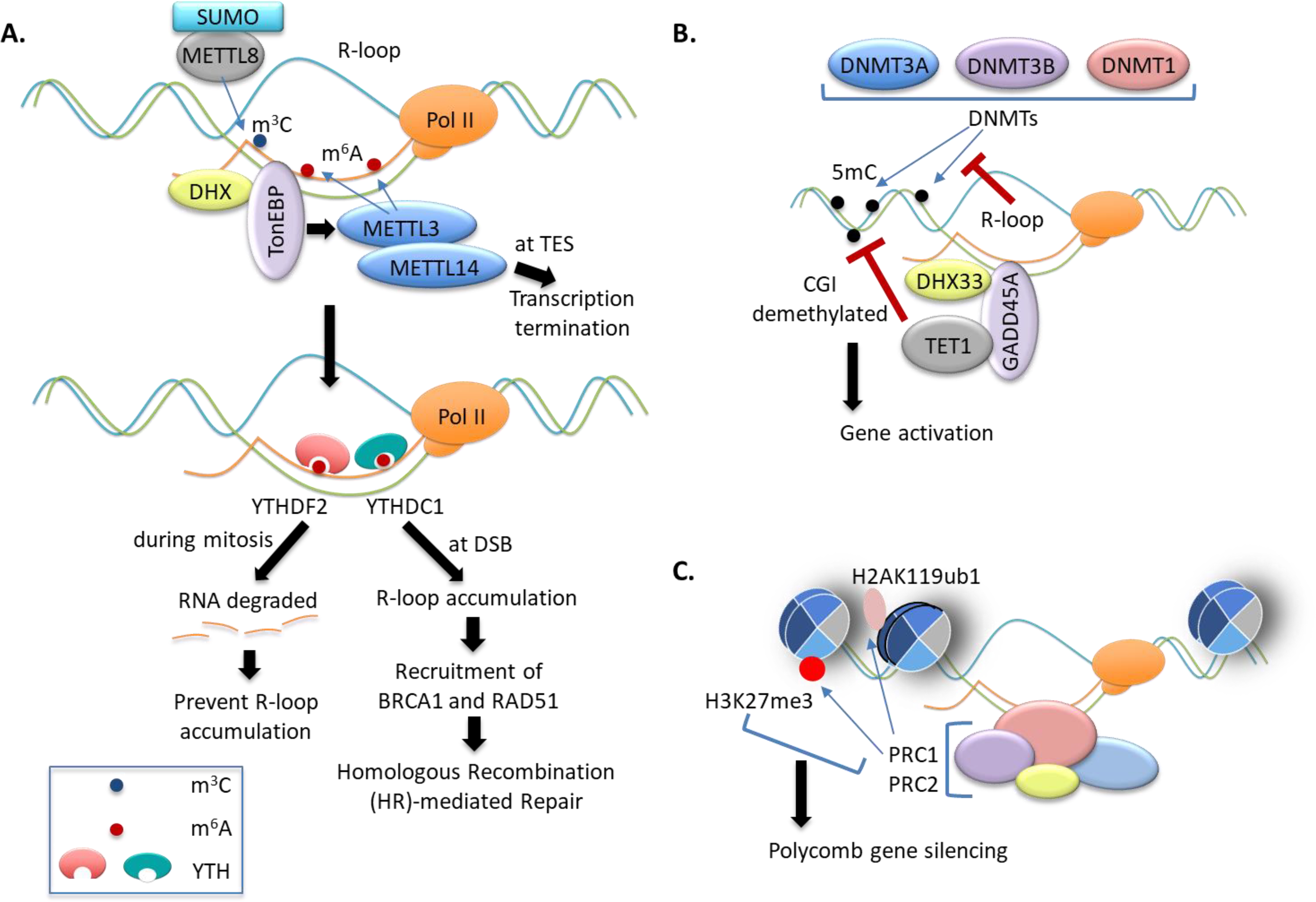
(A) The m6A methylation, installed by METTL3/METTL14 on nascent RNA, or the m3C methylation, installed by SUMOylated METTL8, promotes R-loop formation, which leads to transcription termination at TTSs. The m6A marks in the RNA strands of R-loops can be recognized by m6A “readers” such as YTHDF2 and YTHDC1. In human pluripotent stem cells, YTHDF2 binds to R-loops leading to depletion of m6A-marked R-loops during mitosis. At DSB sites, YTHDC1 increases accumulation of RNA-DNA hybrids and recruits BRCA1 and RAD51 for homologous recombination-mediated repair of DSB.
(B) R-loops are poor substrates for methylation by mammalian DNA methyltransferases (DNMTs). GADD45A binds to R-loop, an event which can be further facilitated by GADD45A interaction with the RNA helicase DHX33, further recruits TET1 to induce CGI demethylation, leading to gene activation.
(C) PcG proteins recognize R-loops and trigger local histone modifications such as H3K27me3 and H2AK119ub1, which provides a mechanism underlying PcG gene silencing.
