Figure 2. Schematic of PEGylated insulin formulations.
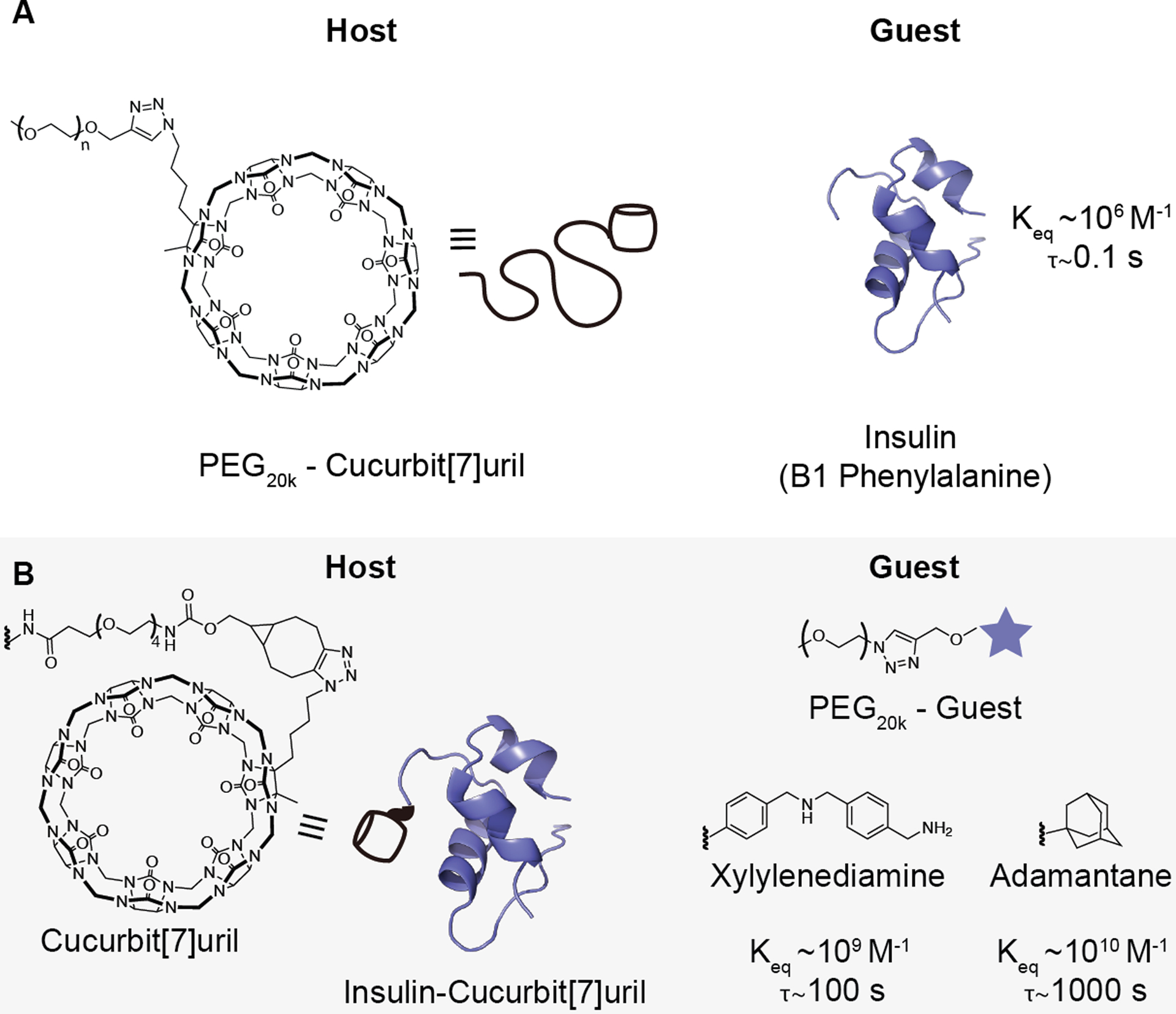
To assess the effect of host–guest binding affinity on the pharmacokinetics of dynamically PEGylated insulin, two non-covalently PEGylated insulin systems were tested. (A) The N-terminal B1 phenylalanine of insulin can bind to cucurbit[7]uril (CB[7]) with a Keq~106 M−1. A PEG20k-CB[7] can thus be used to non-covalently PEGylate the phenylalanine on insulin. (B) To test host–guest motifs with higher binding affinity, a covalent insulin-CB[7] conjugate was combined with a guest-linked PEG20k. To depict the conjugation between CB[7] and insulin, the chemical structures of CB[7], bicyclo[6.1.0]nonyne (BCN), and the PEG linker used are shown (left). PEG20k-xylylenediamine (Keq~109 M−1) or PEG20k-O-adamantane (Keq~1010 M−1) were selected as guests. Bond lifetime (τ), is inversely related to the dissociation rate koff and was calculated based on the following relationships and assumptions: Keq = kon/koff; kon~107; τ = 1/koff. See Table S1 for a complete list of values.
