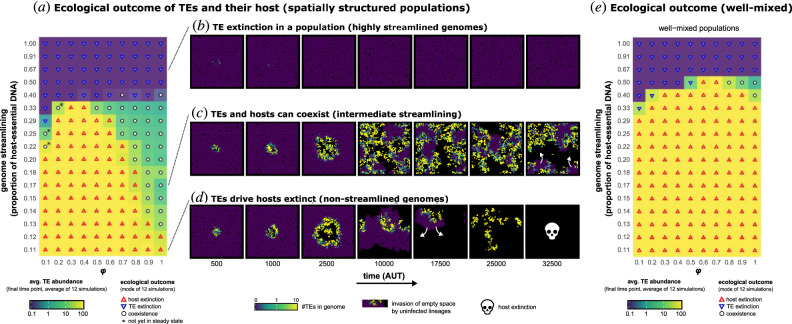
Figure 2.
Spatial structure extends opportunity for coexistence of TEs and hosts at intermediate levels of genome streamlining. (a) Heatmap shows the results of simulations that explore the relationship between the extent of genome streamlining, rate of transposition (φ) and coexistence of TEs and hosts. Simulations marked with an asterisk show apparent coexistence, which after 50 000-time steps have not reached a steady state (see the time courses of TE-abundance for both heatmaps in electronic supplementary material, figure S1). Background colours indicate the average TE abundance in the final time points of the simulations, ranging from zero (purple) to 100 (yellow) on a log scale. (b–d) Visualizations of TE abundance in spatially structured populations where (b) TEs are rapidly lost, (c) TEs stably coexist with their host, or (d) TEs drive the population to extinction. Colours indicate the number of TEs in genomes, ranging from zero (purple) to one (green) to 10 (yellow). (e) Heatmap similar to the one shown in (a), but in well-mixed populations. For this, all individuals are assigned a random position on the grid after each round of replication.