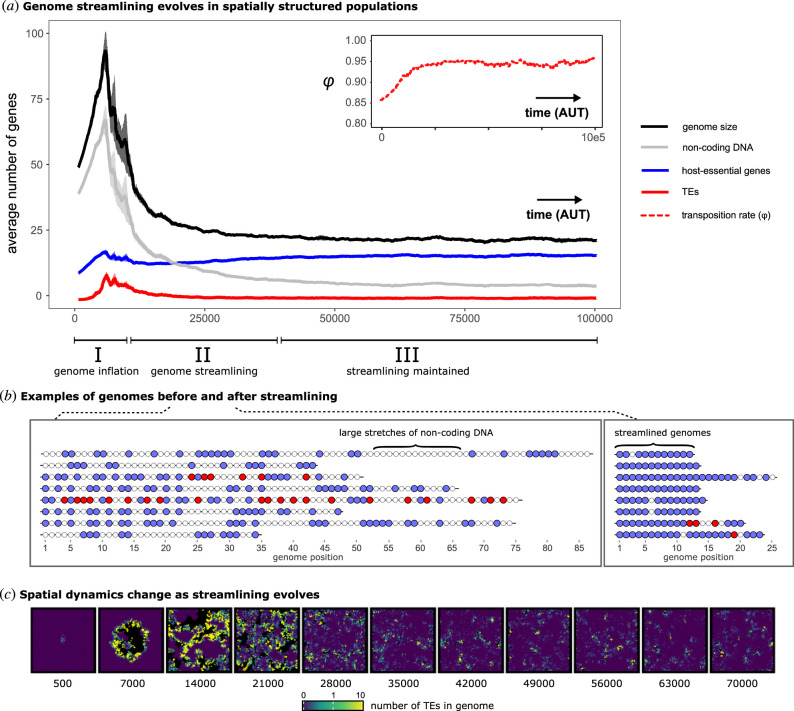
Figure 3.
Coevolution of TEs and host genomes drives genome streamlining. (a) The average number of genes and genome size are plotted over time. Lines show the average of five independent simulations. Shaded areas denote the standard error across simulations. Host-essential genes are shown in blue, TEs are shown in red, and non-coding positions are shown in grey. Note that TEs persist (i.e. the red line is not zero). The inset shows the evolution of the average transposition rate (φ). The ranges of three distinct phases (I, II, and III) in the evolution towards streamlined genomes are shown along the x-axis. (b) Examples of genomes in populations from (a), before and after streamlining has evolved. Blue, red and white dots denote host-essential DNA, TEs and non-coding DNA, respectively. (c) TE-abundance per individual, which correspond to equally spaced time points from a single population from (a). A log-transformed gradient of blue to green to yellow indicates increasing numbers of TEs inside individual genomes. Black indicates empty space where cells have locally died out, which provides niche space for invasion by other lineages.