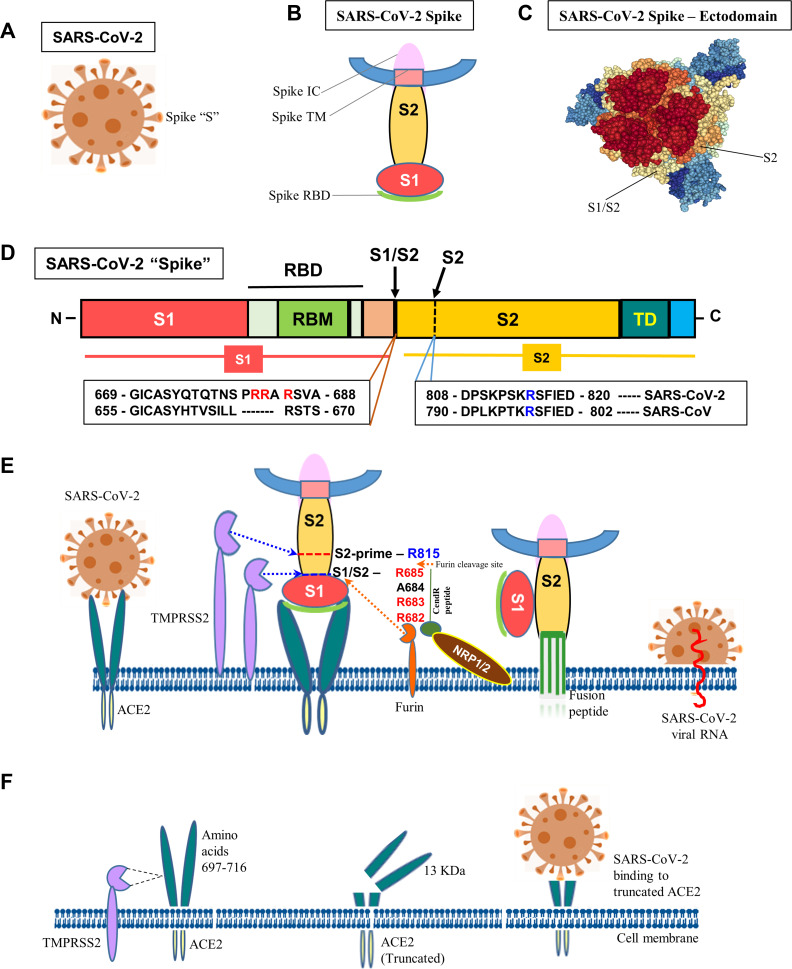
Figure 1.
Molecular mechanism of SARS-CoV-2 infection. (A) Schematic diagram of SARS-CoV-2. (B) “Spike” protein structure and its various operative parts. (C) Cryo-EM structure of “spike” – ectodomain was visualized, S1/S2 junction and S2 cleavage sites were marked by NGL viewer (PDB ID: 7CN9). (D) Schematic diagram of “spike” protein (S) with various functional domains. (E) The diagram shows the entry of SARS-Co-V-2 into a host cell. “Spike” recognizes ACE2 protein and binds to it via the S1 subunit. Subsequently, the S1 is cleaved at the S1/S2 junction with unique amino acid motifs, R682, R683, and R685, or within the S2 subunit by TMPRSS2 and furin that facilitates the structural restriction of S1 on S2. Neuropilin-1 (NRP1) was shown to attach furin-cleaved substrates (S1) in the “C-end rule” CendR motif that potentiates SARS-CoV-2 infectivity. This process releases the internal fusion peptide attached to the spike TM domain for successful viral and cellular membrane fusion. This enables the viral genome to enter the host cell. (F) Diagrammatic representation of TMPRSS2-mediated cleavage of ACE2. The TMPRSS2 could cleave ACE2 at amino acids 697–716 and that results in chopping of a 13 kDa ACE2 fragment and facilitate viral infectivity. The fragment can be easily detected in the cell culture medium.