Figure 3. Phylogenetic analysis of Kirrels.
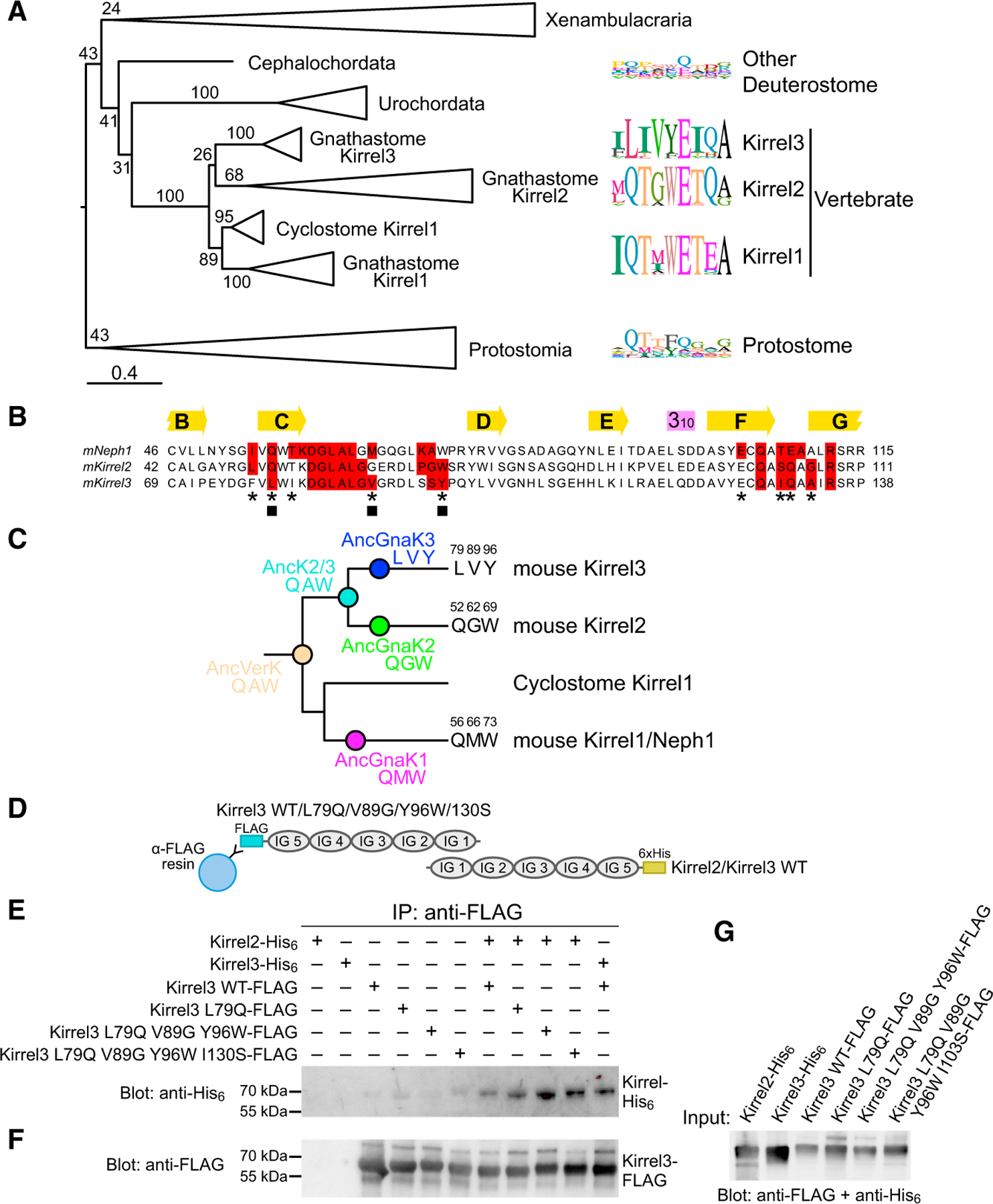
(A) Maximum likelihood tree for Kirrels. The scale bar below represents 0.4 substitutions per site. Numbers on the tree are bootstrap values supporting the adjacent node. The sequence logos to the right show the prevalence of amino acids in selected positions at the Kirrel dimerization interface for specific taxa, placed next to their branch in the phylogenetic tree. Sequence logos were calculated using 26 vertebrate Kirrel1 sequences, 17 vertebrate Kirrel2 sequences, 22 vertebrate Kirrel3 sequences, 8 other deuterostome sequences, and 17 protostome sequences. See Figure S3 for the uncollapsed tree with bootstrap support values.
(B) Sequence alignment showing all amino acids at the interface; red boxes, 4-Å cutoff used for identifying an interface amino acid. The selected residues used in sequence logos in (A) are marked with an asterisk below. Three positions at the interface that vary among ancestral sequences highlighted in (C) are labeled by closed squares.
(C) The three varying residues among sequence reconstructions of the three ancestral gnathostome Kirrels, the Kirrel2/Kirrel3 ancestor, and the ancestral vertebrate Kirrel are shown on the tree. These positions are underlined in the structural views of the interface in Figure 2. See Figure S3 for complete sequences of inferred ancestral D1 domains.
(D) Schematic for the co-immunoprecipitation assay performed between Kirrel2/Kirrel3 wild-type proteins and Kirrel3 wild-type/specificity mutant proteins. FLAG, FLAG-tag; 6×His, hexahistidine tag. WT, wild-type.
(E) FLAG-tagged wild-type and mutant Kirrel3 ectodomains were used to immunoprecipitate hexahistidine-tagged wild-type Kirrel2 or Kirrel3 ectodomains. Only very low levels of wild-type Kirrel2-His6 can be pulled down with wild-type Kirrel3-FLAG; the pull-down becomes increasingly efficient with the Kirrel3 L79Q mutation and the triple and quadruple mutations. For quantitation of the bands in triplicate, see Figure S3C.
(F) The anti-FLAG blot of the samples eluted with FLAG peptide show similar levels of Kirrel3-FLAG captured on anti-FLAG resin in all samples where Kirrel3 FLAG (wild-type or mutant) were included.
(G) The expression levels of His6- and FLAG-tagged Kirrels observed with anti-FLAG and anti-His6 antibodies.
