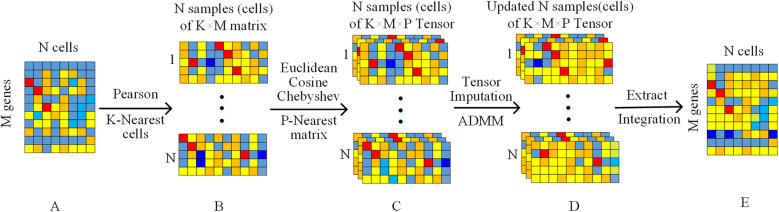
Fig. 11.
The whole framework of scLRTC. For the scRNA-seq dataset A, it uses the PCC and selects the closest K cells to construct N K × M low- rank matrices B. Then it applies the Euclidean, Cosine, and Chebyshev distances to select the closest P low-rank matrices to construct N K × M × P low-rank tensors C. Followingly, it uses the ADMM algorithm to impute the low-rank tensors C to obtain the updated tensors D. Finally, it extracts the cell vector from each low-rank tensor in D and integrates it to obtain the imputed scRNA-seq expression matrix E