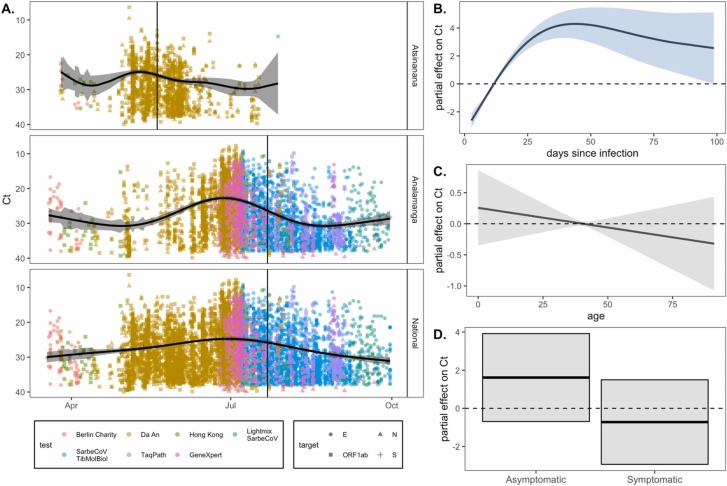
Fig. 2.
RT-qPCR SARS-CoV-2 Ct value as a biomarker of population-level epidemic pace and individual infection trajectory. (A.) Population-level SARS-CoV-2 corrected Ct values from IPM RT-qPCR platform across three Madagascar regions from March-September 2020. Ct values are colored by the test and shaped by the target from which they were derived (legend), though note that all Ct values were first corrected to TaqPath N gene range. The vertical, black line gives the date of peak case counts per region in the IPM dataset, from which these Ct values were derived (May 20, 2020 for Atsinanana and July 22, 2020 for both Analamanga and National). The black, horizontal curve gives the output from a gaussian GAM fit to these data (Table S4), excluding the effects of target and test, which were also included as predictors in the model; 95% confidence intervals by standard error are shown in translucent shading. Partial effects of each predictor are visualized in Fig. S2. Righthand plots visualize partial effects of (B.) days since infection, (C.) patient age, and (D.) patient symptom status on Ct value from our individual trajectory GAM (Table S5). Significant predictors are depicted in light blue and non-significant in gray (Table S5). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)