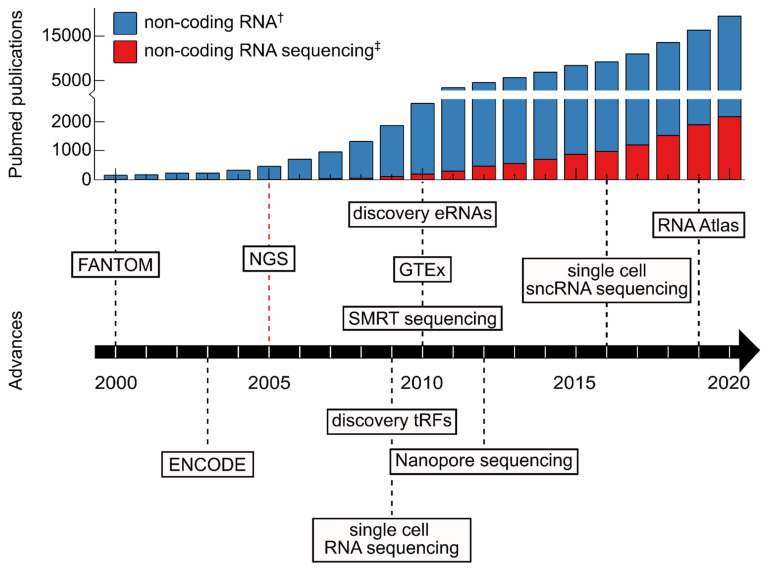
Figure 1.
Important advances in non-coding RNA research over the last two decades. The bar chart presents the numbers of publications with the topics “non-coding RNA” and “non-coding RNA sequencing” per year according to the timeline represented as an arrow. († Pubmed search for “ncRNA” OR “miRNA” OR “piRNA” OR “snoRNA” OR “snRNA” OR “lncRNA” OR “circRNA” OR “non-coding RNA” OR “micro RNA” OR “piwi-interacting RNA” OR “small nucleolar RNA” OR “small nuclear RNA” OR “long non-coding RNA” OR “circular RNA” of 22/10/2021; ‡ Pubmed search for “ncRNA” OR “miRNA” OR “piRNA” OR “snoRNA” OR “snRNA” OR “lncRNA” OR “circRNA” OR “non-coding RNA” OR “micro RNA” OR “piwi-interacting RNA” OR “small nucleolar RNA” OR “small nuclear RNA” OR “long non-coding RNA” OR “circular RNA” AND “sequencing” of 22 October 2021). Selected milestones in non-coding RNA research are displayed on the timeline arrow. The dates for consortia (FANTOM [20], ENCODE [19], GTEx (GTEx Consortium 2013) [25]) were based on the funding year. Advances in sequencing techniques were dated based on their commercial availability (next generation sequencing (NGS) starting with 454 sequencing [18], SMRT sequencing [26], Nanopore sequencing [27]). Novel library preparation methods (single cell RNA sequencing [28], single cell sequencing of small non-coding RNA (sncRNA) [29], databases (RNA Atlas [30]), and newly discovered ncRNA species (tRNA-derived fragments (tRFs) [31], eRNAs [32]) were dated based on their publication years.