Fig. 4 |. Example optical trapping data.
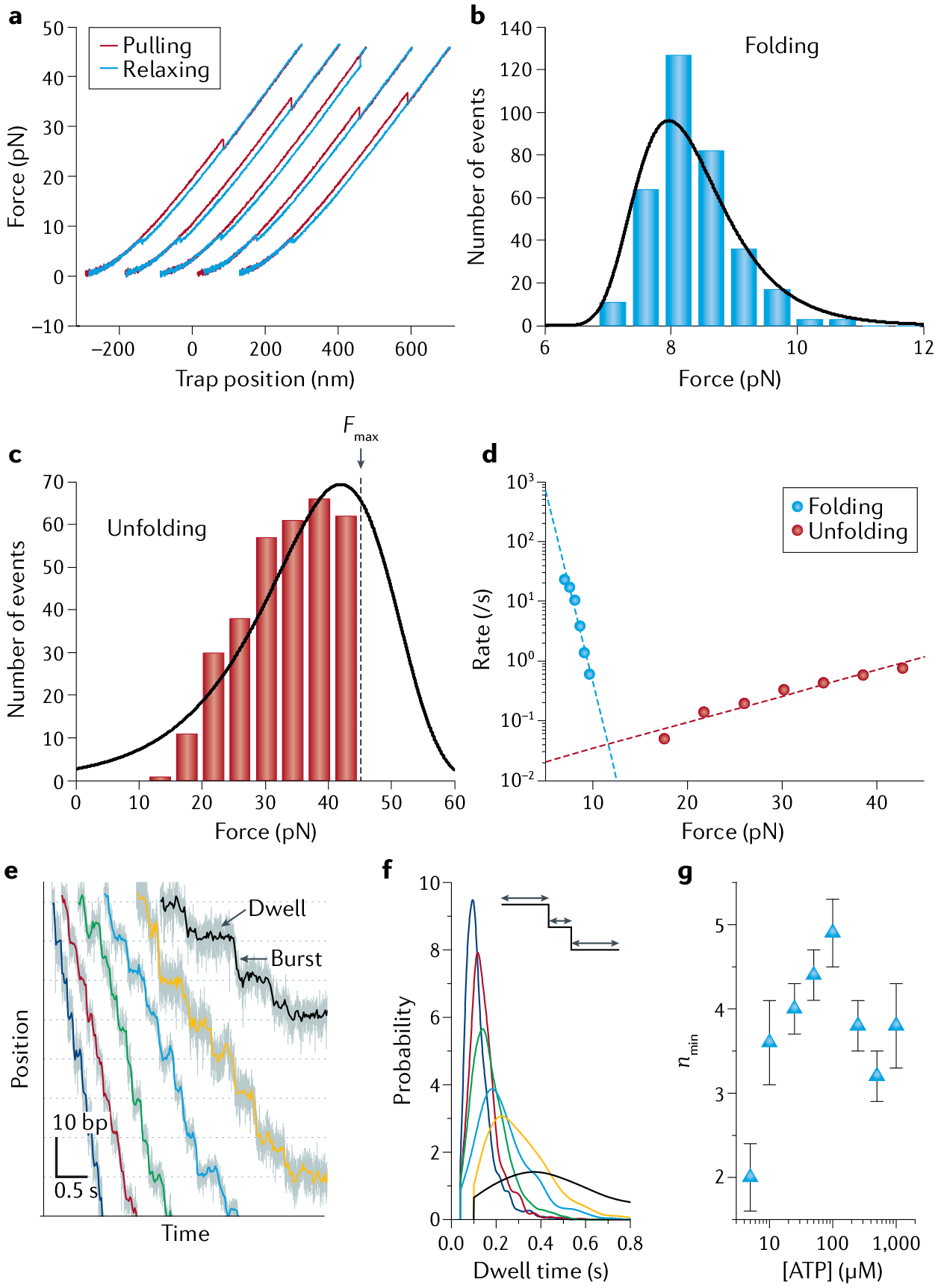
a | Force-ramp cycles of Top7, a de novo designed protein. Successive pulling (red) and relaxing (blue) cycles are offset along the x axis for clarity. b,c | Folding and unfolding force distributions, respectively, of Top7 at a pulling speed of 100 nm/s. Black lines are distributions derived from model fitting in part d. The unfolding distribution in part c is right-censored because the maximum force (Fmax) was set at 45 pN during pulling experiments to avoid tether rupture. d | Force-dependent rates of unfolding (red dots) and refolding (blue dots) extracted from the corresponding force distributions in parts b and c. Dashed lines are fits to Bell’s model254. e | Representative trajectories showing the processive translocation of individual φ29 packaging motors on double-stranded DNA (dsDNA) under a constant force of ~8 pN and different [ATP] (250 μM, 100 μM, 50 μM, 25 μM, 10 μM and 5 μM in blue, red, green, cyan, yellow and black, respectively). Each translocation cycle is composed of a stationary dwell phase and a stepping burst phase. f | Probability distributions of the lifetimes of the dwell phase at different [ATP]. Colour scheme as in part e. g | Values of nmin, the minimum number of rate-limiting kinetic events during the dwell, derived from the dwell time distributions for different [ATP]154. Parts a–d adapted with permission from REF.254, AAAS. Parts e–g adapted from REF.154, Springer Nature Limited.
