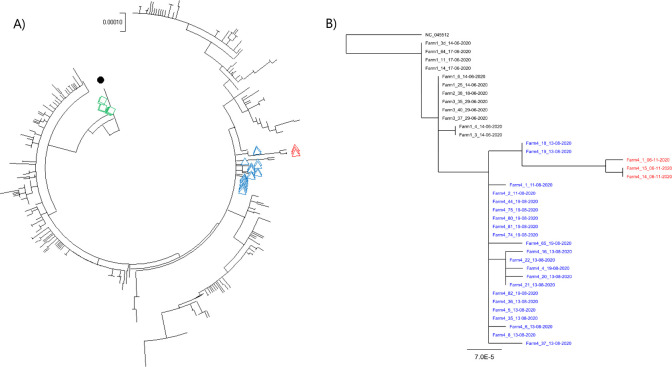
Fig 3. Phylogenetic trees showing the relationships between the full genome sequences of SARS-CoV-2 samples from Danish mink farms with the lineage B.1.1.298 variants.
Panel A. All known SARS-CoV2 lineage B.1.1.298 genome sequences (436 in total) from Danish mink along with the Wuhan reference sequence (NC_045512.2) were included in this analysis. Sequences from the re-infection on Farm 4 (collected in November) are indicated by open red triangles while samples collected in August are indicated by blue triangles. Sequences from Farm 1 are indicated with green squares and the Wuhan reference strain with a black filled circle. The GISAID accession IDs are listed in S1 Table. Panel B. The Maximum Likelihood phylogenetic relationships between the viruses collected on Farm 4 and those from Farms 1–3 are shown. The GISAID accession IDs are listed in S2 Table.