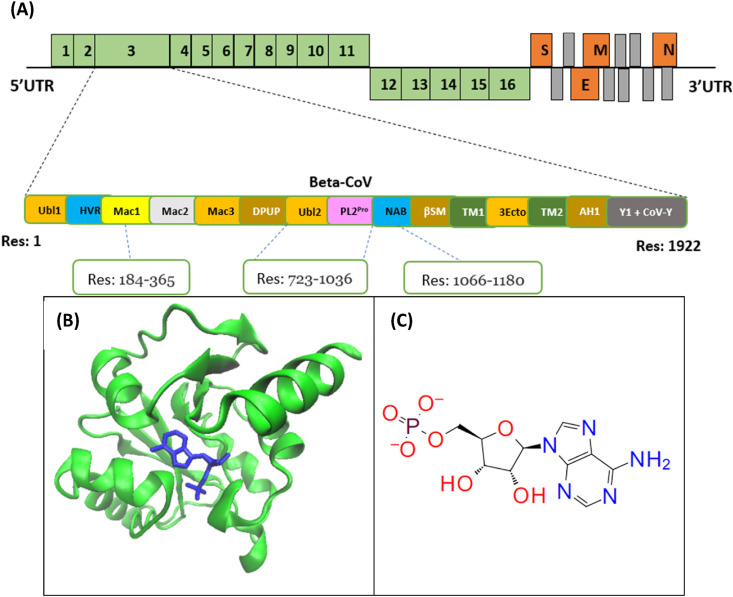
Fig. 1.
(A) Genome organization of Non-structural proteins (green) and structural proteins (orange) of a beta-coronavirus; NSP3 and its domains: Ubl1 (Res: 1–112), HVR (Res: 113–183),Mac1(Res: 184–365), Mac2 (Res: 389–524), Mac3 (Res: 525–652), DPUP (Res: 653–720), Ubl2- PL2Pro(Res: 723–1036), NAB (Res: 1066–1180), βSM (G2M) (Res: 1203–1318), TM1 (Res:1391–1413), 3Ecto (Res:1414–1495), TM2 (Res: 1496–1518), AH1 (Res: 1523–1545), Y1 + CoV–Y (Res: 1546–1922).(B) ADP Ribose phosphatase of NSP3 in complex with AMP (PDB ID: 6W6Y), Residue range of 207–379. (C) Chemical structure of AMP displayed. AMP is used instead of ADP because it is more stable and has the least amount of stored energy.