Fig. 3.
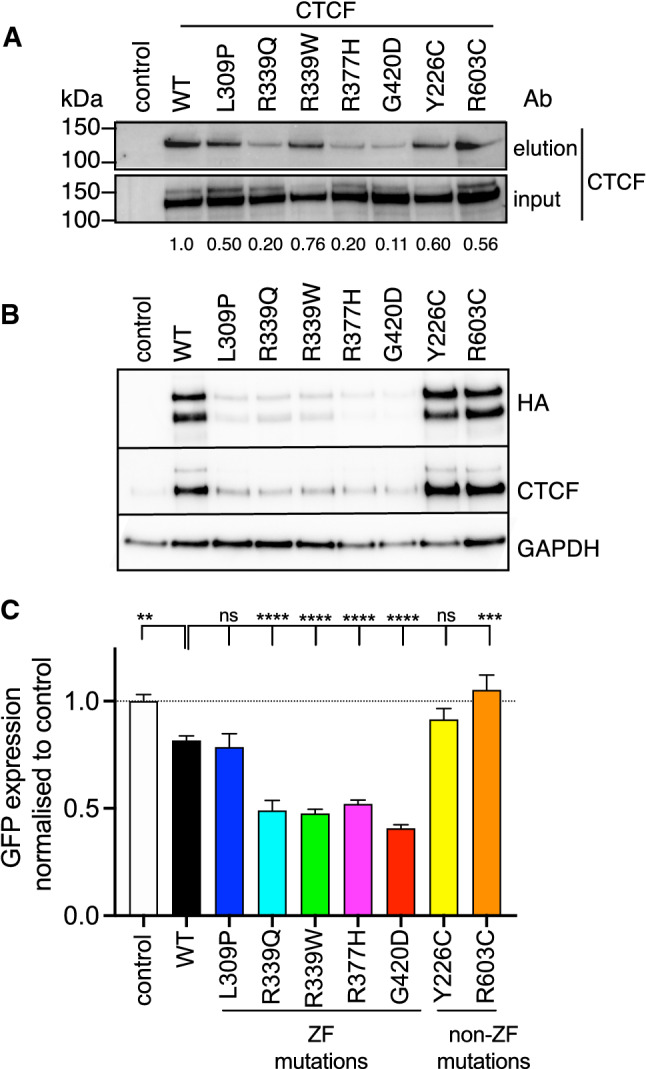
CTCF ZF mutants reduce DNA binding and transcriptional regulation. A CTCF DNA binding assay performed with in vitro transcribed and translated CTCF protein (WT and mutants) and a biotinylated dsDNA probe representing the core CTCF binding site. Eluted and input samples were probed for CTCF protein by Western blot; numbers at the bottom indicate band densitometric values after normalisation to input. B, C Control (eGFP alone), CTCF WT- or mutant-containing lentivector plasmids were transfected into HEK293T cells for 48 h. B Representative Western blots (of 3 replicates) indicating ectopic (HA-tagged) CTCF, total CTCF and GAPDH loading control after transfection of HEK293T cells. C GFP mean fluorescence intensity (MFI) detected after 48 h and normalised to eGFP empty vector control set as 1.0. Data represent the mean ± s.e.m for 3 experiments each performed in triplicate except for the Western blots which are only single replicates. Statistical analysis was performed using a one-way ANOVA with Tukey’s multiple comparisons test for pairwise comparisons between control, WT and mutant (ns = not significant; **p < 0.01; ***p < 0.001; ****p < 0.0001)
