Figure 1. Cartoon depiction of the induced fit, conformational selection, and mixed binding mechanisms.
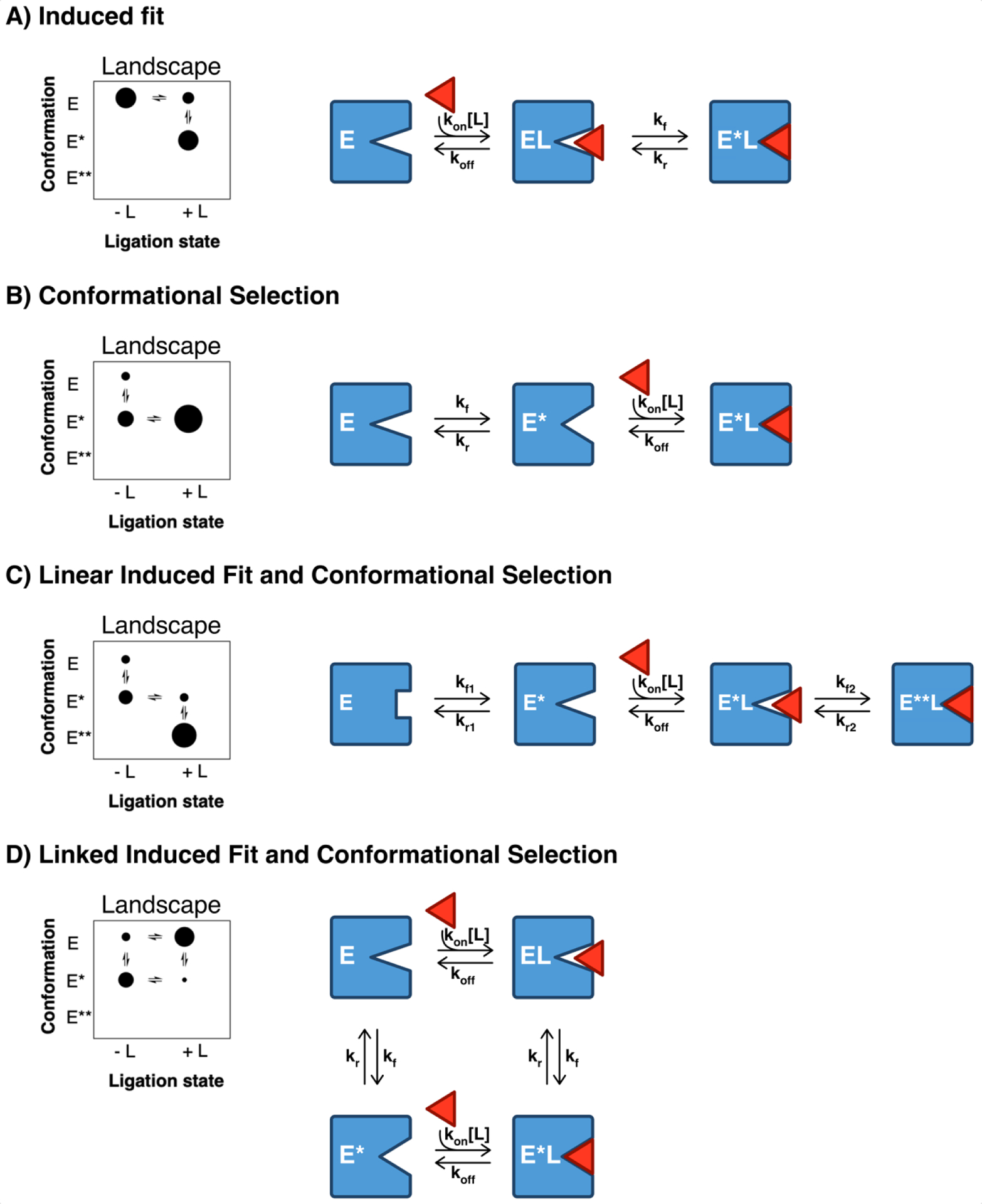
The simplest description of both models contains three enzyme states, and the two models share two common steps, ligand binding and conformational change, the order of which differentiates the models. For IF, ligand binding occurs prior to, and induces, a conformational change, yielding two distinct bound enzyme conformers (EL and E*L in figure 1). Notably, in the pure IF model, the ligand-induced conformer (E*L in figure 1) is only populated upon ligand binding and is otherwise inaccessible in the ligand-free ensemble. CS, on the other hand, occurs when ligand initially encounters an ensemble of unbound states and preferentially binds to (or selects) the ‘binding competent’ conformer (E*), resulting in a population shift to the ‘binding competent’ conformation. The diameter of the dots represents the relative amounts of each enzyme state within each ligation state to illustrate how addition of ligand results in a redistribution of the relative amounts of each enzyme conformer.
