Figure 4: Blood transcriptomic signature of treatment in the LTBI-Other cohort is associated with NK cells, B cells and platelet-related pathways.
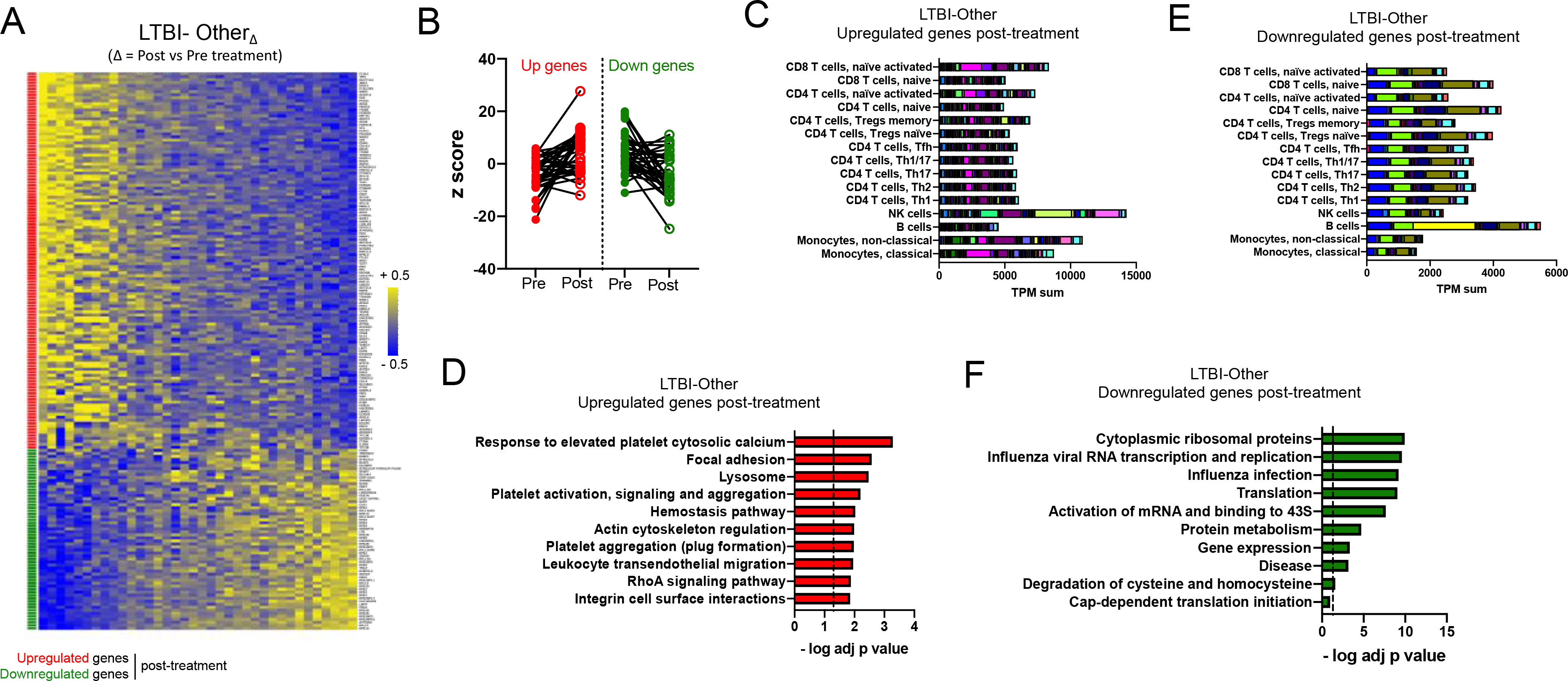
A) Heatmap representing the log fold change expression upon treatment and B) individual z-scores pre and post treatment for the upregulated (red) and downregulated (green) gene-mapped probes in the LTBI-OtherΔ signature derived from differential expression analysis (non-adjusted p value < 0.01, absolute log fold change > 0.2) between post-and pre-treatment paired blood samples in the LTBI-Other cohort (n=36) (Table S6). C) Immune cell type-specific expression and D) top-10 biological pathways enriched in the upregulated genes of the LTBI-OtherΔ signature. E) Immune cell type-specific expression and F) top-10 biological pathways enriched in the downregulated genes of the LTBI-OtherΔ signature. (C,E) For immune cell type-specific expression, each bar consists of stacked sub-bars showing the TPM normalized expression of every gene in corresponding cell type, extracted from the DICE database 34 (http://dice-database.org/). (D,F) Biological pathways were ranked with increasing adjusted p value and dotted line represents significance threshold (adjusted p value < 0.05).
