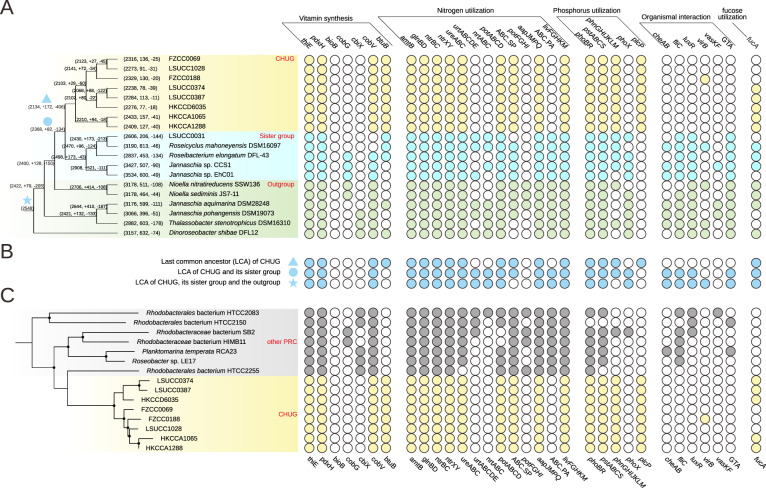
Fig. 4. The phyletic pattern of select genes.
The solid and open circles in the right panel represent the presence/absence of the genes, respectively. A The phyletic pattern in the CHUG, its sister group and its outgroup. The phylogenomic tree shown in the left panel is pruned from the full phylogenomic tree shown in Fig. 1A, and branch length is ignored for better visualization. The ancestral genome reconstruction was performed with BadiRate v1.35 [39]. Each ancestral and leaf node is associated with three numbers, representing the total number of orthologous gene families at this node, and the number of orthologous gene families gained and lost on the branch leading to this node. The last common ancestor (LCA) of CHUG, the LCA shared by CHUG and its sister group, and the LCA shared by CHUG, its sister group and the outgroup are marked with a filled triangle, a filled circle, and a filled star, respectively. B The estimated phyletic pattern of the above-mentioned three LCAs. C The gene presence and absence pattern in the CHUG and other seven PRC genomes. The dendrogram in the left panel is pruned from that shown in Fig. 1B. thiE thiamine-phosphate pyrophosphorylase, pdxH pyridoxamine 5′-phosphate oxidase, bioB biotin synthase, cobG precorrin-3B synthase, cbiX sirohydrochlorin cobaltochelatase, cobV adenosylcobinamide-GDP ribazoletransferase, btuB vitamin B12 transporter, amtB ammonium transport system, glnBD nitrogen regulatory protein P-II, ntrBC nitrogen regulation two-component system, ntrXY nitrogen regulation two-component system, ureABC urease, urtABCDE urea transport system, nrtABC nitrate/nitrite transport system, phoBR two-component phosphate regulatory system, pstABCS phosphate transport system (high affinity), phnGHIJKLM carbon-phosphorus (C-P) lyase, phoX alkaline phosphatase, plcP phospholipase C, cheAB chemotaxis family protein, fliC flagellin, luxR quorum-sensing system regulator, virB type IV secretion system protein, vasKF type VI secretion system protein, GTA gene transfer agent, fucA l-fuculose-phosphate aldolase.