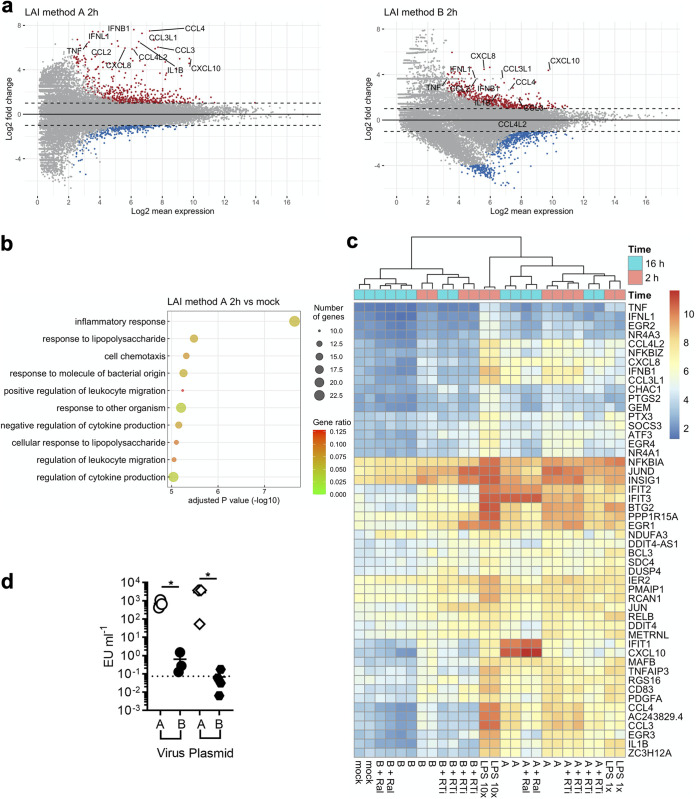
FIG 3.
Virus stocks that activate IFN signaling upregulate LPS-induced genes and contain elevated levels of LPS. (a) Gene expression profiles of two samples of the indicated conditions (cells harvested at 2 h postinfection) were compared to those of mock-infected cells. In these 2-dimensional scatterplots, log2 fold changes between two conditions are used on the y axis, while the log2-transformed mean of normalized expression counts is plotted on the x axis. Shown in red and blue are genes that have a log2 fold change that is greater than 2 or less than –2 and have adjusted P values less than 0.05. Red dots are upregulated genes, while blue dots are downregulated genes. Representative proinflammatory genes are labeled. (b) Top 10 GO terms enriched in the sample from THP-1 cells with virus stock A and harvested at 2 h postinfection. See the legend of Fig. 2 for details. (c) Transcriptional profiles of THP-1 cells infected with HIV-1 under various conditions were examined based on significantly upregulated genes by LPS (10×, 200 μg/mL; 1×, 20 μg/mL). The top 50 genes with largest variance among LPS-induced genes across these samples were chosen to draw the heat map using a normalized read count. Abbreviations are the same as those used in Fig. 2. Time points of sample harvesting are shown by colored boxes on the top of the heat map. Samples collected at 2 and 16 h postinfection are shown in magenta and cyan, respectively. (d) The amount of LPS in plasmid and virus stocks was measured with a standard assay using amebocyte lysates. Data values were generated using four different plasmid stocks and three virus stocks. Data sets were analyzed using the two-tailed, unpaired Student’s t test. A horizontal dashed line represents the limit of detection, which is the lowest value within the linear range of the standard curve that falls in the linear range. EU, endotoxin unit; *, P < 0.05.