Figure 2.
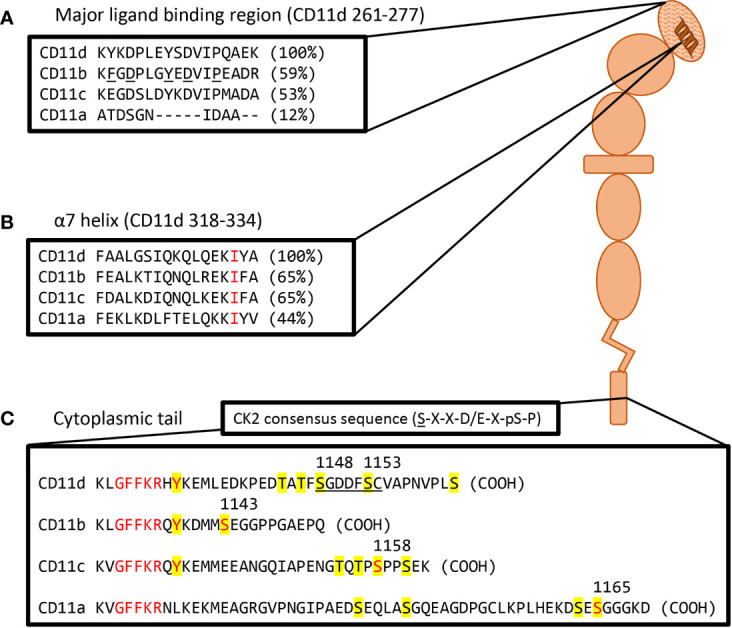
Visual representation of a probable CD11d structure including amino acid homolog comparisons of key motifs. (A) Sequence comparison of the CD11d α-I domain major ligand binding region. The ligand binding CD11d α-I domain is highlighted by a hatched pattern. Residues determined to be important in the ligand binding pocket of CD11b are underlined and percent homology to CD11d is in brackets. Alignment and CD11b residue analysis performed in previous study (51). (B) Sequence comparison of the CD11 α7 helix. An invariable isoleucine is highlighted in red and percent homology to CD11d is in brackets. Alignment was performed in previous study (17). Conformational changes to the α7 helix within CD11d have been shown to alter ligand affinities, thus implying the presence of an open and closed α-I domain conformation (51). (C) Sequence comparison of complete CD11 cytoplasmic tails. Yellow denotes potential phosphorylation sites, red denotes conserved residues of interest, and the underlined sequence denotes a potential CK2 site. The GFFKR “hinge” motif is required to maintain the association of the CD11 and CD18 cytoplasmic tails (52). The constitutive phosphorylation of a serine residue is conserved across CD11a (Ser1165) (53), CD11b (Ser1143) (54), and CD11c (Ser1158) (55). CD11d has a putative CK2 site at Ser1148-Cys1154 using the consensus sequence (S-X-X-D/E-X-pS-P) (56). The same sequence would predict Ser1153 to be constitutively phosphorylated as observed in other β2 integrins. The function of the conserved GFFKR +2 tyrosine residue in CD11b-d is largely undefined. The tyrosine appears to be embedded into the membrane during the inactive conformation, while exposed during the active conformation (57). Long isoform CD11a (NP_002200.2), CD11b (NP_001139280.1), CD11c (NP_000878.2), and CD11d (NP_001305114.1) amino acid sequences were acquired from the National Center for Biotechnology Information database (28–31).
