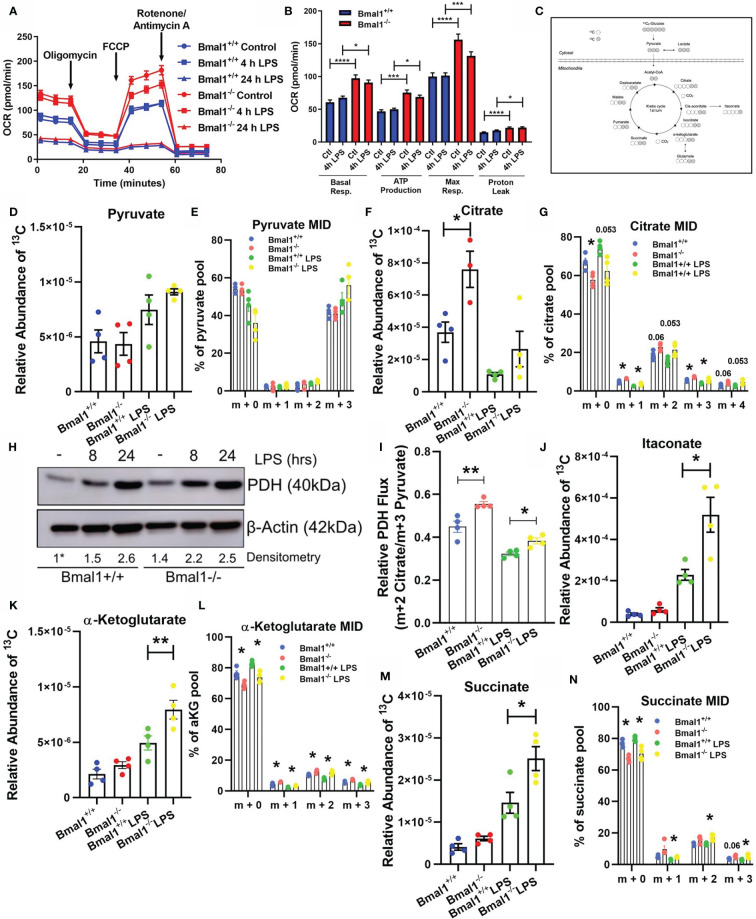
Figure 1.
Mitochondrial respiration and Krebs cycle glucose flux is altered in Bmal1-/- macrophages. Bmal1+/+ and Bmal1-/- BMDMs were stimulated with LPS (100 ng/ml), subjected to a Seahorse XF mitochondrial stress test. (A) OCR of BMDMs was measured, and the metabolic trace illustrates changes in OCR following injection of the mitochondrial stress test compounds oligomycin, FCCP, and rotenone/antimycin a. (B) Measures of basal respiration, ATP production, maximal respiration, and proton leak were derived from (A). Assay results are presented +/- SEM and are representative of n=3 independent experiments. BMDMs were isolated, seeded in 10 mM U-13C6 glucose, and stimulated with LPS for 8 hours. BMDMs were lysed and metabolites were quenched and measured via GC-MS to trace Krebs cycle flux of labelled glucose. (C) A schematic of U-13C6 glucose-derived carbon Krebs cycle flux and incorporation into metabolic intermediates. Relative abundance of U-13C6-labelled (D) pyruvate, (F) citrate, (J) itaconate, (K) α-Ketoglutarate, and (M) succinate, and mass isotopologue distribution (MID) of (E) pyruvate, (G) citrate, and (L) α-Ketoglutarate, and (N) succinate were measured. MID values of low abundance isotopologues are excluded for clarity. (I) Ratio of m+2 citrate/m+3 pyruvate representative of U-13C6 glucose-derived carbon flux through pyruvate dehydrogenase. Data presented is n=4 +/- SEM. (H) BMDMs were stimulated with LPS and protein expression of PDH was analysed by Western blot using β-Actin as a loading control. Densitometry is relative to the Bmal1+/+ control band. This band is indicated by a * symbol. Data presented is representative of n=3 independent experiments. Statistical analysis was performed for Seahorse XF data and U-13C6 relative abundance values by one-way ANOVA with Sidak’s multiple comparisons test and for MID values by multiple student’s t-tests with Holm-Sidak correction for multiple comparisons (*p < 0.05, **p < 0.01 ***p < 0.001, ****p < 0.0001).