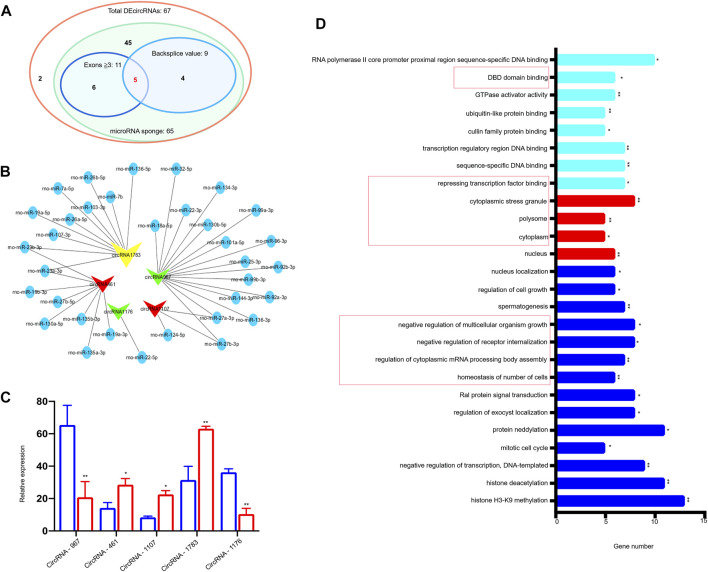
FIGURE 4.
Characterization of 5 DEcircRNAs in rat subcutaneous adipose tissue. (A) Venn show abundant (exons ≥3) exonic DEcircRNAs with potential miRNA binding sites and DEcircRNAs with backsplice value of every sample. (B) Bioinformatics prediction of 5 circRNAs, their predicted miRNAs, and miRNA targets. circRNAs were plotted as V-shapes. Yellow V-shapes represent circRNA-1783 (circ-ATXN2), which is an up-regulated circRNA. Green V-shapes represent down-regulated circRNAs (circRNA-967, circRNA-1176). Red V-shapes represent up-regulated circRNAs (circRNA-461, circRNA-1107). miRNAs were plotted as blue nodes. (C) Expression pattern of the 5 DEcircRNAs between young and old rat subcutaneous adipose tissue based on mRNA sequencing. *p < 0.05, **p < 0.01 compared with young group. (D) GO analysis of the 5 DEcircRNAs. Using GO database (http://www.geneontology.org) analysis the GO enrichment of DEcircRNAs, based on three aspects: biological processes (BP, blue strip), cellular components (CC, red strip), and molecular functions (MF, sky blue strip). GO terms which in the red frame were related to the circRNA-1783 (circ-ATXN2). *represent the significance of the GO term enrichment among the 5 selected DEcircRNAs. *p < 0.05, **p < 0.01 compared with young group. Expression was normalized to actin and expressed as mean ± SEM (n = 3), by two-way ANOVA.