FIGURE 1.
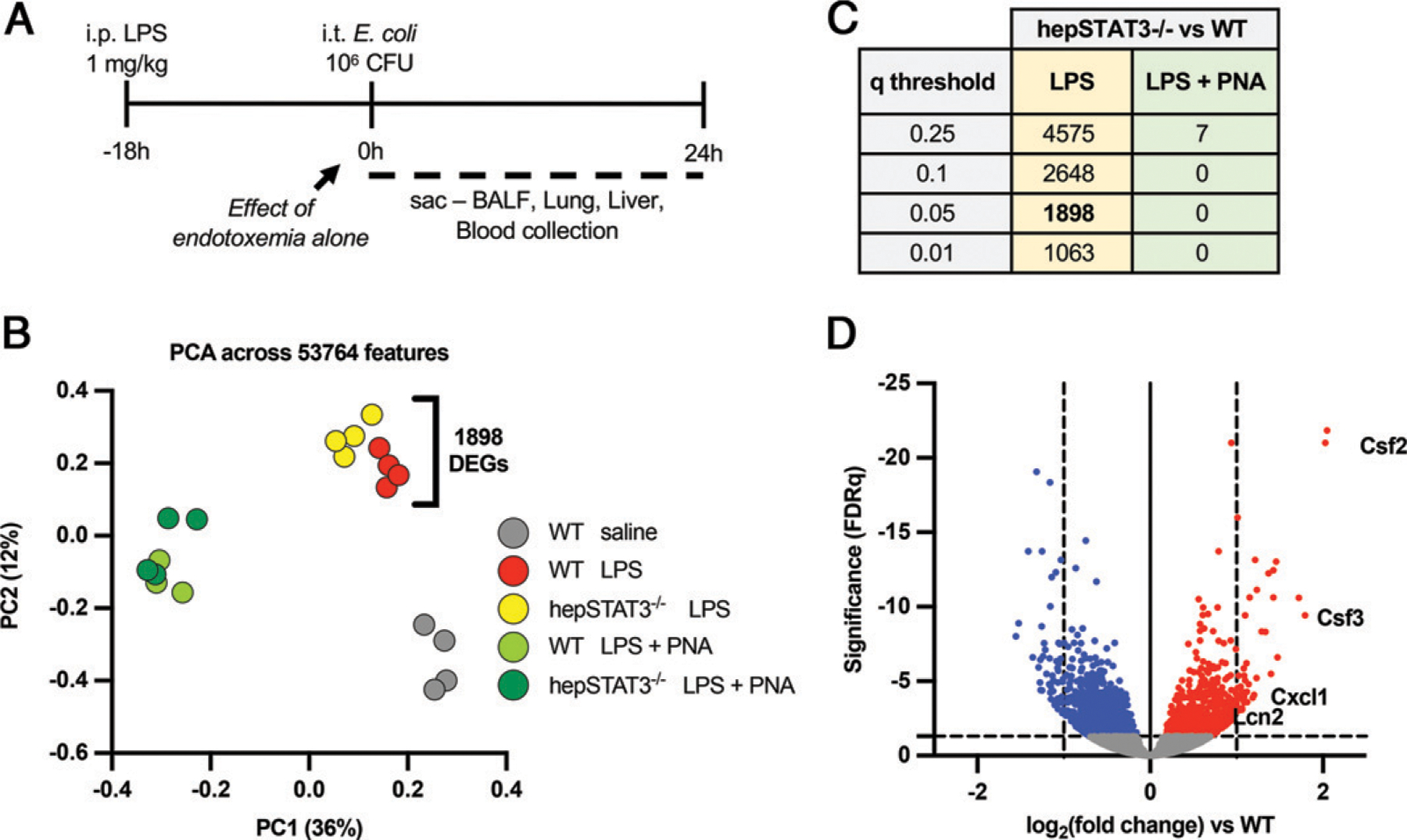
Liver activity modifies the lung transcriptome during endotoxemia in a genotype-dependent manner. (A) Two-hit model of endotoxemia and pneumonia. WT and hepSTAT3−/− mice received i.p. LPS (1 mg/kg) for 18 h and were either collected then (at 0 h) or given a direct pulmonary infection via an i.t. instillation of E. coli (106 CFUs) for an additional 24 h. (B) RNAseq was performed on WT or hepSTAT3−/− mice challenged with LPS for 18 h and either 0 or 24 h of pneumonia (PNA). A control group of WT mice with 18 h i.p. saline was included. The principal component analysis (PCA) plot indicates a distinct genotypic clustering between WT and hepSTAT3−/− mice after endotoxemia alone, with 1898 significant gene changes between the lungs of each group. (C) The table indicates the number of DEGs between hepSTAT3−/− and WT lungs at 18 h i.p. LPS and at 18 h LPS/24 h pneumonia with the corresponding q value. (D) The volcano plot depicts genes in hepSTAT3−/− lungs that are significantly (q < 0.05) increased (red) or decreased (blue) versus WT controls after 18 h i.p. LPS. n = 3–4/group from four independent experiments.
