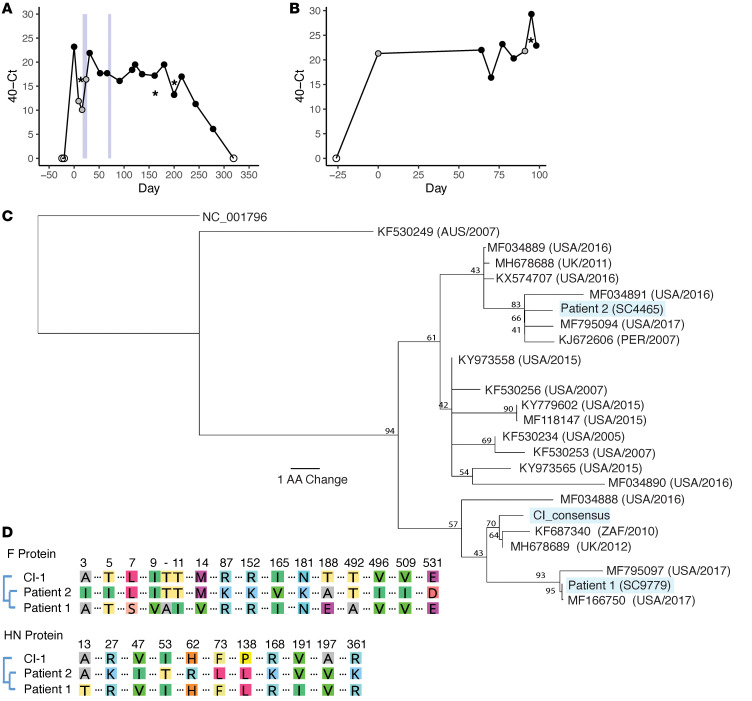
Figure 1. Longitudinal sampling of long-term HPIV3 infections in vivo.
Sampling time series data with associated Ct values shown for patient 1 (A) and patient 2 (B). Samples collected by nasal swabs are represented by dots; black dots are HPIV3-positive samples with associated sequencing data, gray dots are HPIV3-positive samples that were not available for sequencing, and empty dots tested negative for HPIV3. HPIV3-positive bronchoalveolar lavage (BAL) samples are represented by asterisks. Light blue boxes indicate time periods in which patient 1 was treated with DAS181. Amino acid sequences of the HN protein for patient 1 and patient 2 are placed in the context of circulating strains that were downloaded from NCBI’s GenBank (C). All sequences are labeled with GenBank accession number followed by collection location and collection date. Consensus support values are shown next to branch points. The laboratory-adapted HPIV3 reference strain (NC_001796) is used as an outgroup. Amino acid alignment of the day 0 consensus sequences of the HN and F attachment proteins for each patient were aligned with the clinical isolate used in functional assays (D). Only amino acids that differ among these isolates are depicted.