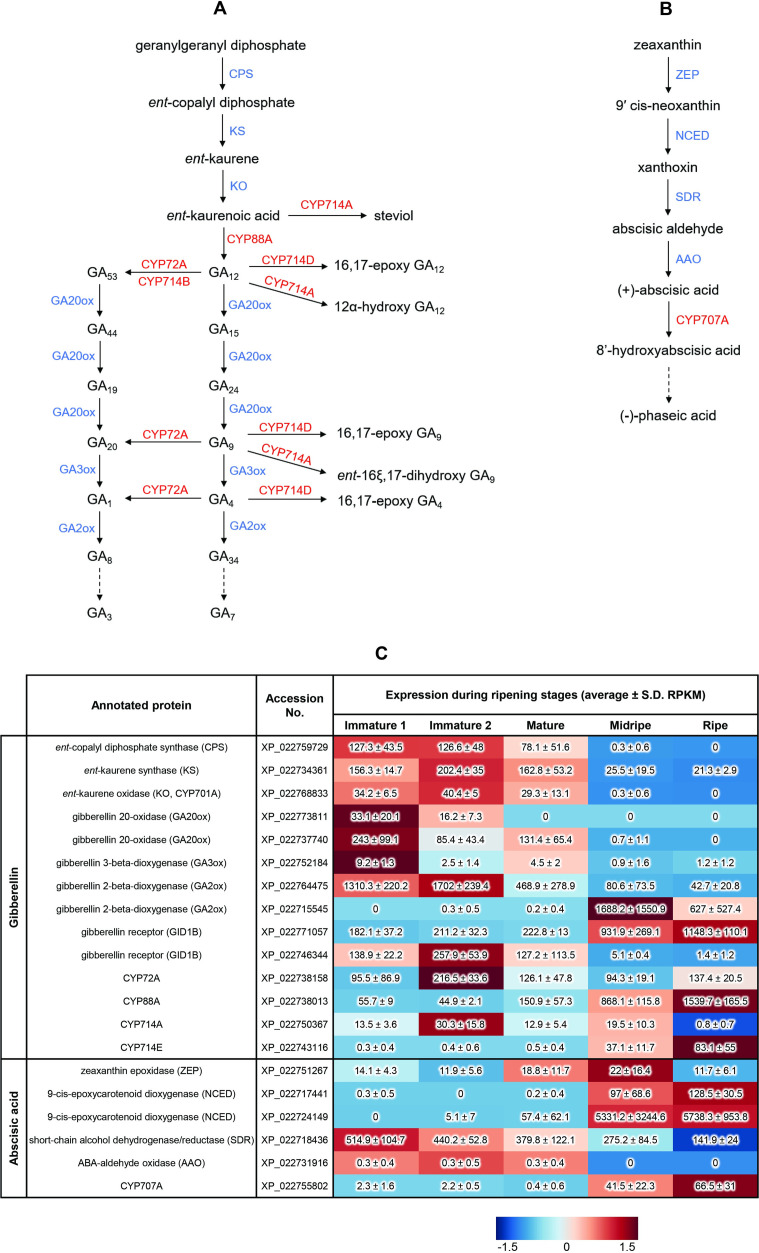
Fig 5. Proposed reaction and RNA-seq based expression profiles involved in phytohormone biosynthesis.
(A) Gibberellin biosynthetic pathway driven by CYP72A, CYP88A, and CYP714 in durian. Enzyme abbreviations: CPS, ent-copalyl diphosphate synthase; KS, ent-kaurene synthase; KO, ent-kaurene oxidase; GA2ox, gibberellin 2-oxidase; GA3ox, gibberellin 3-oxidase; GA20ox, gibberellin 20-β-oxidase. Cytochrome P450s are indicated in red, whereas other enzymes are indicated in blue. (B) Proposed catalytic pathway of abscisic acid driven by CYP707A in durian. Enzyme abbreviations: ZEP, zeaxanthin epoxidase; NCED, 9-cis-epoxycarotenoid dioxygenase; SDR, short-chain alcohol dehydrogenase/reductase; AAO, ABA-aldehyde oxidase. Cytochrome P450s are indicated in red, whereas other enzymes are indicated in blue. (C) Expression profiles of putative genes involved in the biosynthesis of gibberellin and abscisic acid during the ripening stages of the aril of the Monthong cultivar. The average RPKM values with standard deviations are shown in the table. The heatmap represent RPKM values, generated by MetaboAnalyst 5.0. Higher expression for each gene is presented in red; otherwise, blue was used.