Figure 1.
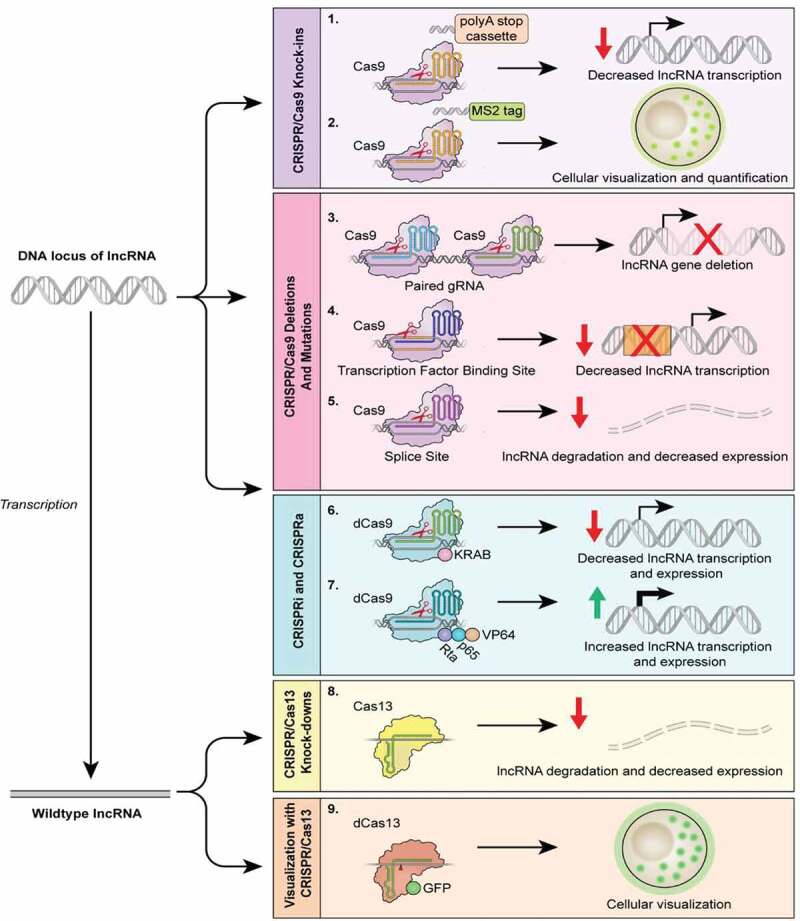
CRISPR uses in studies of lncRNAs. The CRISPR/Cas9 technology can be used to target various regions within a lncRNA gene, manipulating the lncRNA expression to gain functional insights. The CRISPR/Cas9 technology can be used to produce knock-in phenotypes (1,2), inserting a polyA stop cassette downstream of the transcription start site to decrease transcription or to encode a MS2 tag, as used in the CERTIS system, to localize lncRNAs. Additionally, the Cas9-gRNA complex can be directed to the beginning and end of the locus (3), splice sites (5), as well as transcription factor binding sites within the promoter region (4). These techniques will likely result in the partial or complete reduction of lncRNA levels within a cell. CRISPR activation, CRISPRa (7), and CRISPR interference, CRISPRi (6), can be used to overexpress or silence lncRNA expression via catalytically dead Cas9 (dCas9) without permanent genomic editing. CRISPRa employs a dCas9-gRNA complex fused with transcription activators (i.e. VP64, p65, and Rta) to robustly activate the transcription of the lncRNA gene. Conversely, the transcription of a lncRNA gene can be inhibited by CRISPRi, which utilizes a dCas9-gRNA complex fused with KRAB, a transcription repressor. Various orthologs of Cas13 can be utilized in lncRNA knockdown (8) in which a gRNA targets catalytically active Cas13 to single-stranded lncRNAs, leading to degradation of the molecule. Cas13 can also be used to visualize lncRNAs (9) by fusing dCas13 to a fluorescent protein
