Figure 1.
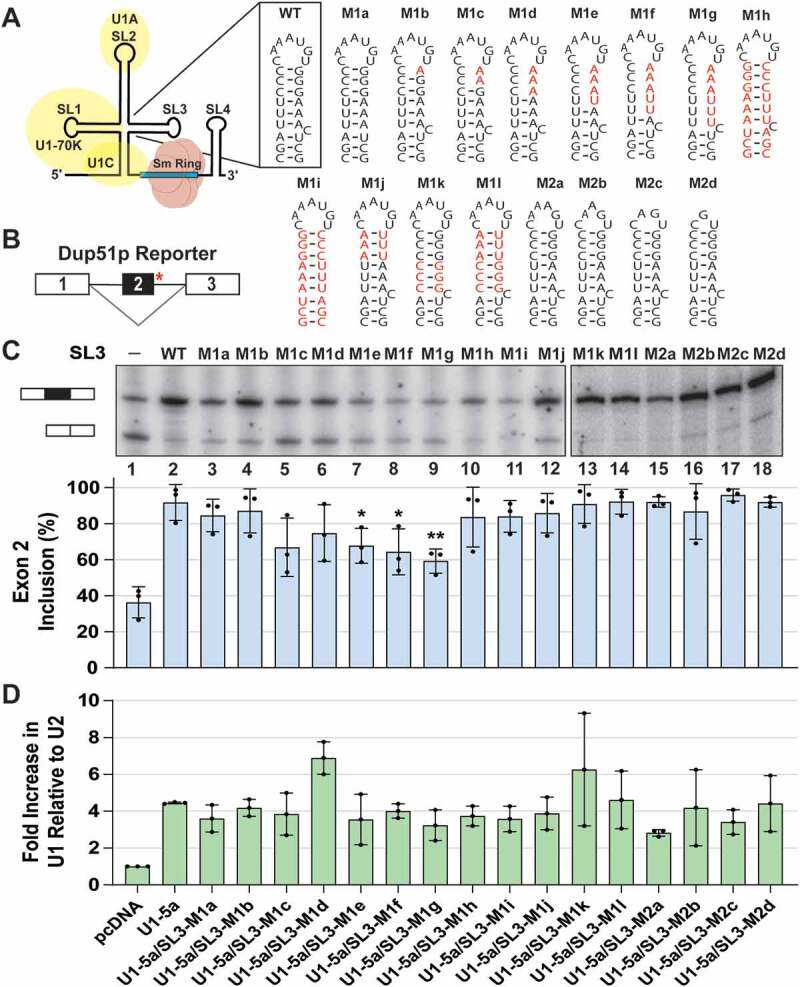
Stem-loop 3 of the U1 snRNA is important for U1 function. (A) Schematic diagram of the U1 snRNP. Secondary structure and sequence of wildtype SL3 and mutations introduced into the U1 snRNA are depicted; nucleotide changes are shown in red. (B) Dup51p pre-mRNA carries a 5′-ss mutation (indicated by the red asterisk) in intron 2 that causes skipping of exon 2 in the mature transcript. (C) Primer extension analysis to monitor splicing of the minigene reporter Dup51p after cotransfections with control (pcDNA) or U1-5a plasmids expressing wildtype or mutant U1 snRNAs. The full-length and exon 2 skipped Dup51p mRNA products are depicted. The percentage of the full-length product (± s.d., n = 3) is represented in the graph below and statistical significance was determined by comparisons to the wildtype control (lane 2) (n = 3; * = p < 0.05, ** = p < 0.01). (D) RT-qPCR analysis of U1 snRNA expression in HeLa cells cotransfected with Dup51p and U1-5a variants carrying wildtype SL3 or mutations. Fold change in U1 snRNA expression was calculated relative to the pcDNA control after normalization to U2; fold change (± s.d.; n = 3) in U1 is graphed. Expression of the Dup51p reporter pre-mRNA upon cotransfection with U1-5a/SL3 mutants M1e, M1f, and M1g (lanes 7–9) appears to be reduced in this experiment. This apparent diminution, however, is not a consistent observation, as the transcript level is not reduced in Fig. 2B, lane 4
