Figure 3.
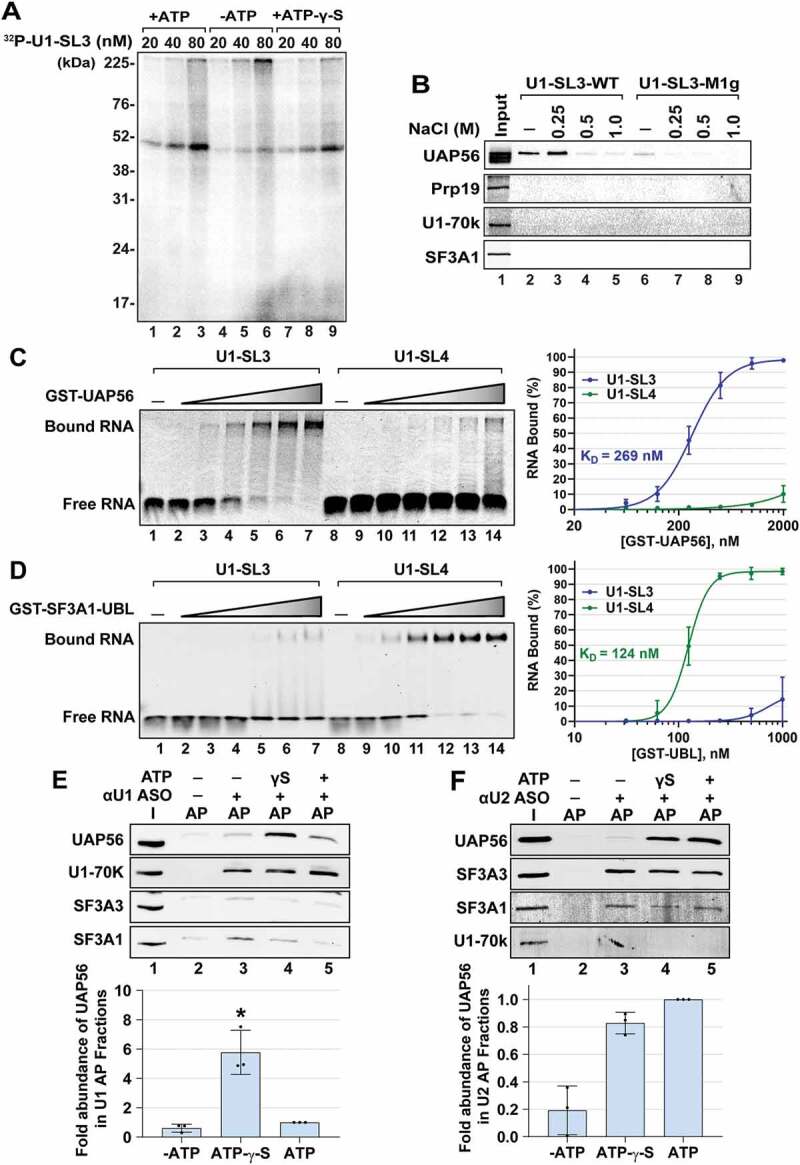
SL3 and SL4 bind to distinct spliceosomal proteins. (A) UV crosslinking analysis for U1-SL3 interacting protein(s). HeLa nuclear extracts were incubated with 20, 40, and 80 nM 32P-U1-SL3 RNA in the presence or absence of ATP and ATP-γ-S. (B) Western analysis of proteins in wildtype and mutant U1-SL3 complexes. HeLa nuclear extracts were preincubated with 0, 0.25, 0.5, and 1.0 M NaCl prior to RNA affinity purification using biotinylated U1-SL3-WT and U1-SL3-M1g RNAs. (C) EMSAs monitoring binding of Cy5-labelled U1-SL3 (lanes 1–7) or U1-SL4 (lanes 8–14) RNAs (10 nM) in the absence and presence of GST-UAP56 (0.0625, 0.125 0.25, 0.5, 1.0, and 2.0 μM). (D) EMSAs monitoring binding of Cy5-labelled U1-SL3 (lanes 1–7) or U1-SL4 (lanes 8–14) RNAs (10 nM) to GST-SF3A1-UBL (0.03125, 0.0625, 0.125 0.25, 0.5, 1.0 μM). Displayed dose-response curves were generated by plotting the average percent of bound U1-SL3 and U1-SL4 RNA (± s.d., n = 3) versus GST-UAP56 or GST-SF3A1-UBL protein concentration and the apparent affinity constant values (KD) are reported. (E) Western analysis of proteins present in input (I) and U1 affinity purified (AP) complexes in the absence and presence of ATP-γ-S, and ATP. The intensity of the UAP56 band was normalized to that of U1-70k in the U1 and U2 complexes, and then fold change was calculated relative to the plus ATP condition (± s.d., n = 3, * = p < 0.05). (F) Western analysis of proteins present in input (I) and U2 AP complexes in the absence and presence of ATP-γ-S, and ATP. Because the signal for SF3A3 was better than SF3A1, the intensity of UAP56 band was normalized to that of SF3A3 protein in the U2 complexes, respectively, and then change was calculated relative to the plus ATP condition (± s.d., n = 3, * = p < 0.05)
