Figure 3.
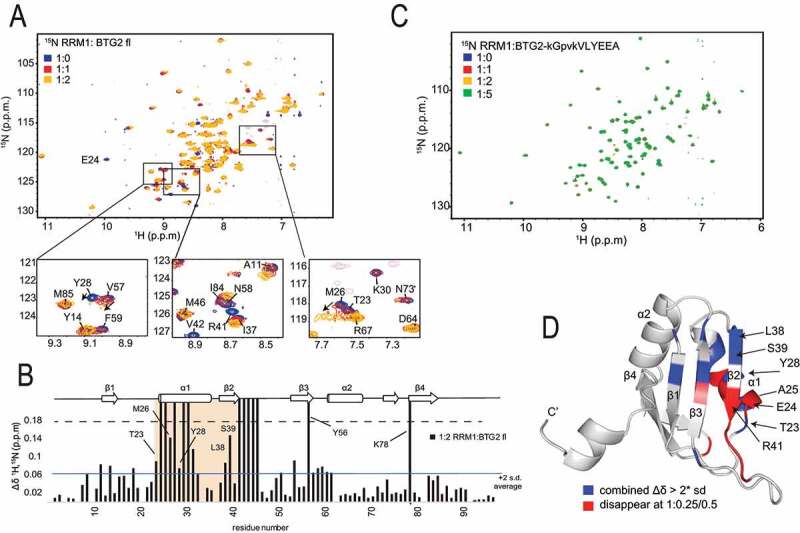
NMR identification of the PABPC1 RRM1 residues involved in the interaction with BTG2
A) Overlay of 1H-15N HSQC spectra of free RRM1 (blue) and in complex with increasing amounts of BTG2 (1:1 red; 1:2 orange). Some key residues are indicated by a box and the close-up view shown below. In the close-up view, arrows show the direction of the chemical shift change. Aliased negative peaks in pink correspond to the amides of arginine side chains. The number of scans (NS) is 2, otherwise indicated if different. B) The combined chemical-shift perturbations of RRM1 upon binding to BTG2 (1:2.5 ratio; blue). The secondary structure is represented at the top and the region with high chemical shift changes (in α1 andβ 2) is highlighted in orange. Key residues are labelled. The horizontal line represents two times the s.d. of all chemical shift differences. Peaks which disappear at 1:0.25 or 1:0.5 and did not return at higher ratios are shown by bars with >0.18 ppm (dashed line). C) Overlay of 1H-15N HSQC spectra of RRM1 (blue) and RRM1 in complex with increasing amounts of BTG-kGpvkVLYEEA (1:1 red; 1:2 yellow; 1:5 green). D) Cartoon representation of RRM1; modified from PABP RRM12 structure (pdb code 1CVJ). Chemical-shift perturbations upon BTG2 addition larger than 2× s.d. are coloured in red while signals which disappear at 1:0.25/0.5 are shown in blue
