Figure 1.
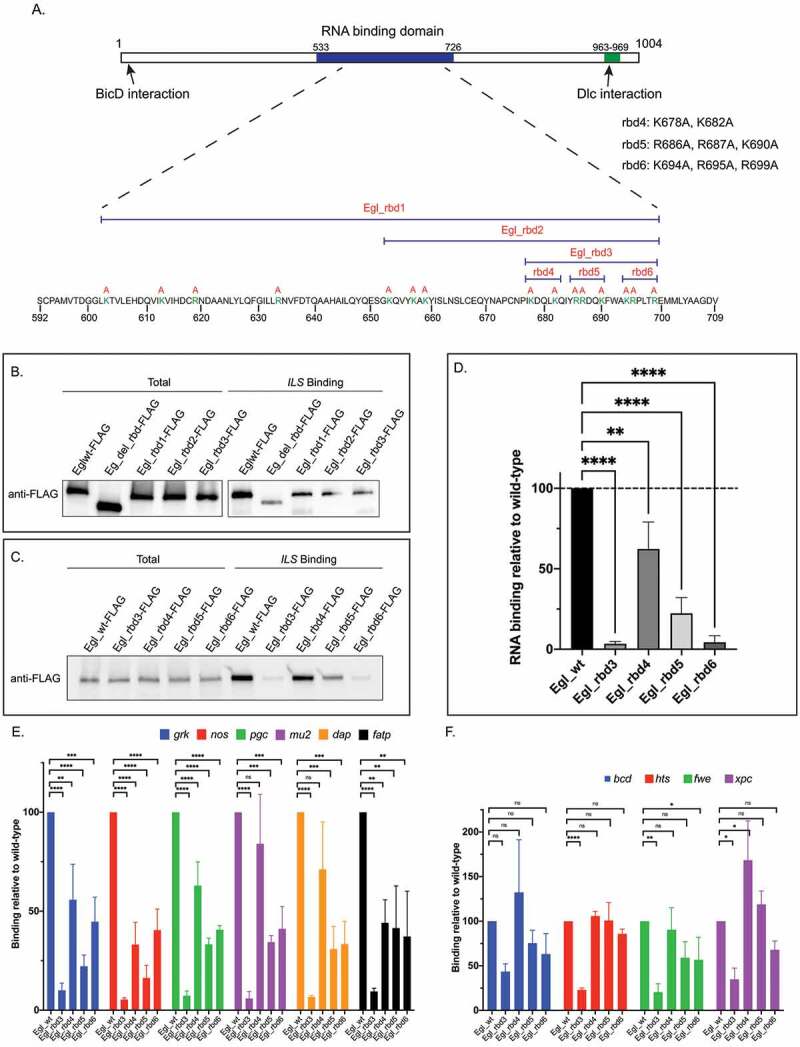
Identification of Egl RNA binding mutants. (a) Schematic showing the domain structure of Egl. The RNA binding domain, and the BicD and Dlc interaction sites are shown. The positively charged residues that were mutated are shown in green. The specific residues mutated in Egl_rbd4, Egl_4bd5 and Egl_rbd6 are also shown. (b) The indicated Flag-tagged Egl constructs were expressed in S2 cells. Cell lysates were incubated with Streptavidin beads coated with the ILS localization element. After binding and wash steps, the bounds proteins were eluted and analysed by western blotting using the FLAG antibody. The total fraction was also analysed. (c) A similar RNA binding experiment was performed using ovarian lysates from flies expressing the indicated Egl constructs. Bound proteins were eluted and analysed by blotting using the FLAG antibody. A total fraction is also shown. (d) Quantification of the experiment shown in C. The experiment was repeated three times and the band intensity in the bound fractions was determined. Values are shown relative to the wild-type construct. (e-f) Ovarian lysates were prepared from flies expressing the indicated Egl constructs. The FLAG-tagged proteins were immunoprecipitated using FLAG beads. The co-precipitating RNAs were extracted, reverse transcribed and analysed using quantitative PCR. RNA enrichment was normalized to the level of gamma tubulin mRNA that co-precipitated in each pellet. The values are shown relative to binding observed with wild-type Egl. ****p ≤ 0.0001, ***p ≤ 0.001, ** p ≤ 0.01, *p ≤ 0.05, ns = not significant; a one-way ANOVA was used to calculate statistical significance
