Figure 3.
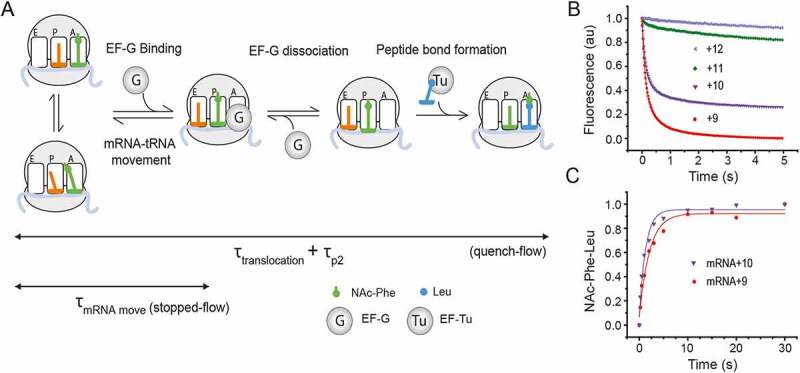
Kinetics of translocation of the pre-T complex containing NAc-Phe-tRNAPhe and pyrene labelled mRNAs
(A) Schematic representation of the peptide elongation cycle on the ribosome starting from the pre-equilibrated pre-T complexes, which are in equilibrium between the classical and the hybrid state. EF-G binding and GTP hydrolysis lead to mRNA movement by one codon. The ribosome complex undergoes certain structural rearrangements and EF-G releases, thereby leaving the ribosome ready for the next peptide bond formation. The meantime of the whole elongation cycle (τtranslocation + τp2) was determined by rapidly mixing NAc-Phe-tRNAPhe containing pre-T complex (0.5 µM) with EF-G (5 µM) and Leu TC (1 µM) in a quench-flow and following the formation of NAc-Phe-Leu against time. In parallel, the kinetics of the mRNA movement was followed in stopped-flow, where EF-G (5 µM) was rapidly mixed to a NAc-Phe-tRNAPhe containing pre-T complex (0.5 µM). As a result of the mRNA movement, the fluorescence of the 3ʹ pyrene labelled mRNAs decreases, which is monitored in stopped-flow against time. The τmRNA move was determined from the reciprocal of the rates obtained from the fluorescence traces.(B) Kinetics of mRNA movement upon EF-G binding to the pre-T complexes containing NAc-Phe-tRNAPhe and pyrene labelled mRNAs: mRNA+9 (red circle), mRNA+10 (violet inverted triangle), mRNA+11 (green diamond) and mRNA+12 (light purple left triangle). The pyrene fluorescence was monitored with 360 nm long-pass filter (343 nm excitation).(C) Kinetics of NAc-Phe-Leu formation in a quench-flow by rapid mixing of a pre-T complex with EF-G and Leu TC.
