Figure 1.
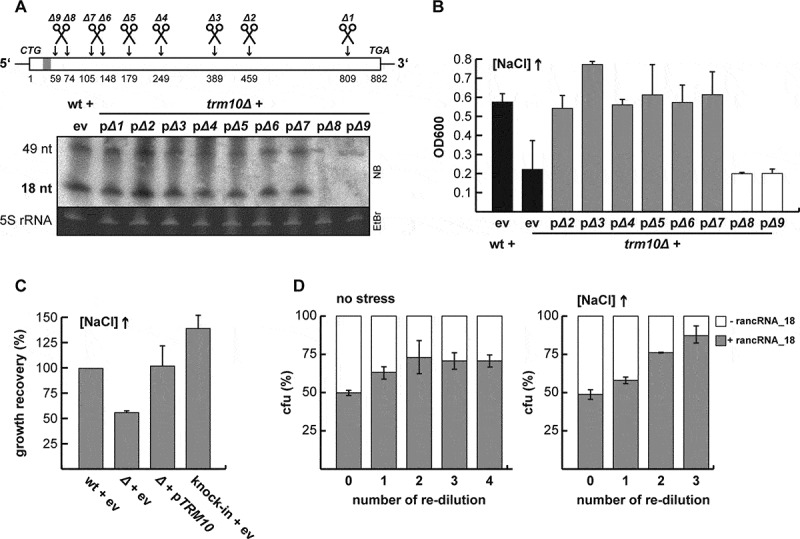
rancRNA_18 is a bona fide ncRNA in S. cerevisiae. (A) schematic overview of the truncated versions (Δ1 – Δ9) of the TRM10 locus. the grey bar close to the 5ʹ end depicts the location of rancRNA_18. the presence of rancRNA_18, and the putative 49-mer processing intermediate, was assessed via northern blot analysis (5S rRNA served as loading control). all constructs were expressed from a plasmid in the trm10Δ strain, whereas the original start codon was replaced by a CTG codon to prevent translation of the transcripts (11). ev, empty vector control. (B) growth recovery after 1,050 minutes under hyper-osmotic conditions of wildtype (wt) and trm10Δ cells (black bars) was compared to trm10Δ cells expressing truncated versions of the TRM10 locus. n = 2, mean ± SD. (C) growth of the knock-in strain that expresses the truncated TRM10 construct Δ6 from a novel genomic location (knock-in + ev) in high salt media was compared to the wt cells or to the strain carrying either no additional plasmid (ev) or the plasmid encoding the full-length TRM10 locus (pTRM10). n = 6, mean ± SD. (D), growth competition between a mixture of equal starting amounts of two strains differing only in the absence (white) or presence (grey) of rancRNA_18 expression in normal (no stress) or high salt containing media. cultures were grown to stationary phase and colony forming units (cfu) determined. cultures were then re-diluted and cfu determined at each stationary phase. n = 3, mean ± SD
