Figure 3.
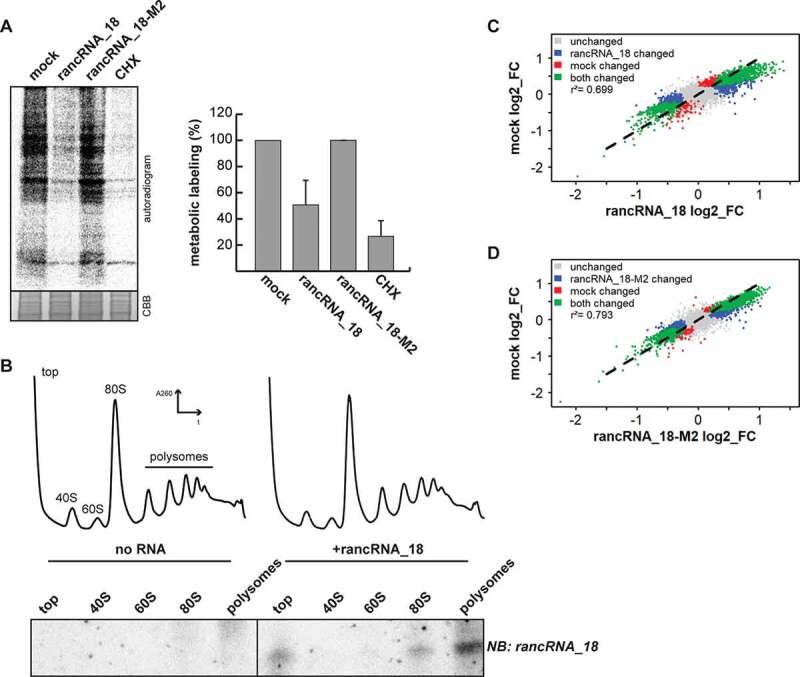
rancRNA_18 is a global translational inhibitor. (A) spheroplasts from trm10Δ cells were electroporated with synthetic rancRNA_18 or the mutant version rancRNA-18-M2 and used for metabolic labelling. electroporation in the absence of synthetic RNA (mock) and samples containing the translational inhibitor cycloheximide (CHX) served as controls. radioactively labelled proteins were analysed with SDS-PAGE and autoradiography. coomassie brilliant blue (CBB) stained proteins serve as loading control. signals were quantified and normalized to the no RNA (mock) control. n = 2, mean ± SD. (B) polysome profiling was performed with recovered spheroplasts, RNA was isolated from collected fractions and separated on a denaturing polyacrylamide gel. the distribution of the synthetic rancRNA_18 in the retrieved fractions was visualized by northern blot analysis. (C and D) scatter plots showing translation efficiencies (log2 fold-changes; FC) of sequencing data from total (T) and polysome-associated (P) mRNA from spheroplasts electroporated with no RNA (mock), synthetic rancRNA_18 or rancRNA_18-M2, respectively. different colours indicate mRNAs that show a different association to polysomes in both conditions (green), in samples electroporated with rancRNA_18 only or rancRNA_18-M2 only (blue), in mock samples only (red), or in none of the conditions (grey). p-values for control vs rancRNA_18 data and control vs rancRNA_18-M2 data were <2.2 •10−16. correlation coefficients are indicated as r2
