Figure 4.
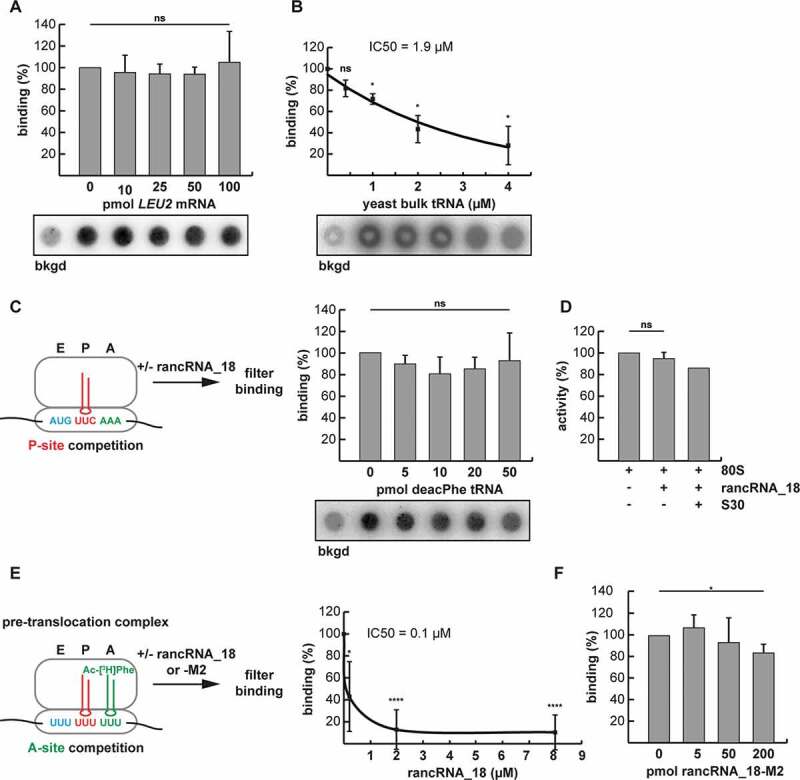
RancRNA_18 has an influence on A-site tRNA binding. (A) dot blot filter binding analysis was used to analyse binding competition of radiolabeled rancRNA_18 with increasing amounts of LEU2 mRNA to 5 pmol purified yeast ribosomes. the signal in the absence of 80S particles (bkgd) was subtracted. ns, not significant. n = 2; mean ± SD. (B) as in (A) but with increasing amounts of yeast bulk tRNA. The rancRNA_18 binding data (n = 3; mean ± SD; * p < 0.05) are blotted as function of increasing tRNA competitor concentration and the trend-line is depicted. (C) dot blot approach to investigate ribosome binding competition between radiolabeled rancRNA_18 and P-site bound deacylated tRNAPhe (red). n = 3; mean ± SD. (D) peptidyl transfer between P-site bound N-acetyl-[3H]Phe-tRNAPhe and A-site located puromycin was assessed in the absence or presence of rancRNA_18. S30 cell extract was used as a control if additional factors are needed for rancRNA_18 functionality. n = 2; mean ± SD. (E) A pre-translocation complex formed on poly(U) mRNA analogues carrying deacylated tRNAPhe in the P-site (red) and N-acetyl-[3H]Phe-tRNAPhe in the A-site (green) was used to investigate effects of unlabelled rancRNA_18 addition on tRNA occupancy. extend bound A-site tRNA was assessed via filter binding. n = 6; mean ± SD; * p < 0.05, **** p < 0.0001. (F) as in (E) but in the presence of increasing amounts of rancRNA_18-M2
