Figure 2.
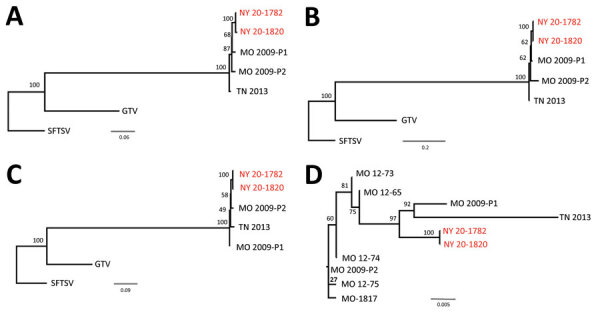
Phylogenetic relationship among Heartland virus isolates, Suffolk County, Long Island, New York, USA. Separate alignments of large segments (A), medium segments (B), small segments (C), and partial nonstructural sequences (D) were created with MAFFT in Geneious version 11.1.5 (https://www.geneious.com). Maximum–likelihood analyses were completed with RAxML (https://cme.h–its.org) using 1,000 bootstraps. Bootstrap values are indicated at each node. Phylogenetic trees for each segment were rooted to SFTSV strain HB154 (GenBank accession nos. JQ733560–62). Guerta virus strain DXM was included as an additional outgroup (GenBank accession nos. 328591–93). New York isolates from this study (red text), together with the 3 previously available full–genome sequences (MO 2009–P1 [patient 1, GenBank accession nos. JX005842, 4, 6]; MO 2009–P2 [patient 2, GenBank accession nos. X005843, 5, and 7]; and TN 2013 [TN, GenBank accession nos. J740146–8]), were included in these analyses (panels A, B, and C). Six additional partial sequences available for a 606–nt region of the nonstructural protein gene (GenBank accession nos. C466555, KC466560, KC466561, KC466562, KC466563, and MT052710) are indicated in an unrooted maximum–likelihood tree in panel D. Scale bars indicate nucleotide substitutions per site. GTV; Guerta virus; SFTSV, severe fever with thrombocytopenia syndrome virus.
