Figure 1.
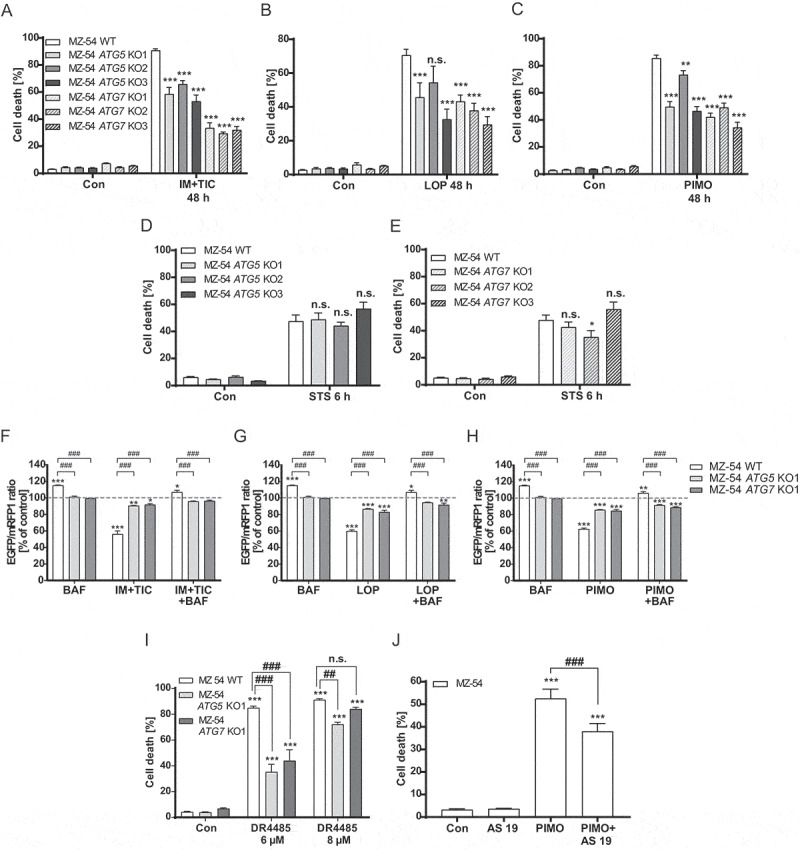
Determination of autophagic cell death upon treatment with loperamide and pimozide. (A-E) Flow cytometric analysis of cell death by measurement of APC-ANXA5 binding and PI uptake. MZ-54 WT cells and three different CRISPR-Cas9 ATG5 and ATG7 KO cell lines were treated with 20 µM imipramine + 100 µM ticlopidine (IM+TIC) (A), 12.5 µM loperamide (LOP) (B), 12.5 µM pimozide (PIMO) (C) for 48 h or with 3 µM staurosporine (STS) (D and E) for 6 h. A-E display total cell death including only-APC-ANXA5-positive, only-PI-positive and double-positive cells. DMSO was used as vehicle (Con, 48 h or 6 h, respectively). Data show mean + SEM of at least three experiments with three replicates and 5,000–10,000 cells measured in each sample. Statistical significances were calculated with a two-way ANOVA. (F-H) Flow cytometric analysis of the autophagic flux by measurement of the EGFP and mRFP1 signal. MZ-54 WT cells and ATG5 and ATG7 KO cells were exposed to 20 µM imipramine + 100 µM ticlopidine (IM+TIC) (F), 15 µM loperamide (LOP) (G) or 15 µM pimozide (PIMO) (H) for 16 h alone or in combination with bafilomycin A1 (BAF). Data show mean + SEM of three experiments with three replicates and 5,000–10,000 cells measured in each sample. Statistical significances were calculated with a two-way ANOVA. (I and J) Assessment of cell death as described above. (I) DR4485 was added in a concentration of 6 µM or 8 µM to MZ-54 WT cells or the respective ATG5 and ATG7 KOs. DMSO (con) served as control. Data represent mean + SEM of three independent experiments with three replicates and 5,000–10,000 cells measured in each sample. Statistical significances were calculated with a two-way ANOVA. (J) 10 µM AS 19 was added 2 h before MZ-54 WT cells were treated with 12.5 µM pimozide (PIMO) for 40 h. DMSO (con) served as control. Data represent mean + SEM of four independent experiments with three replicates and 5,000–10,000 cells measured in each sample. Statistical significances were calculated with a one-way ANOVA Significances between WT and KO cells are depicted as P ≤ 0.05: #, P ≤ 0.01: ## or P ≤ 0.001: ###
