Figure 1.
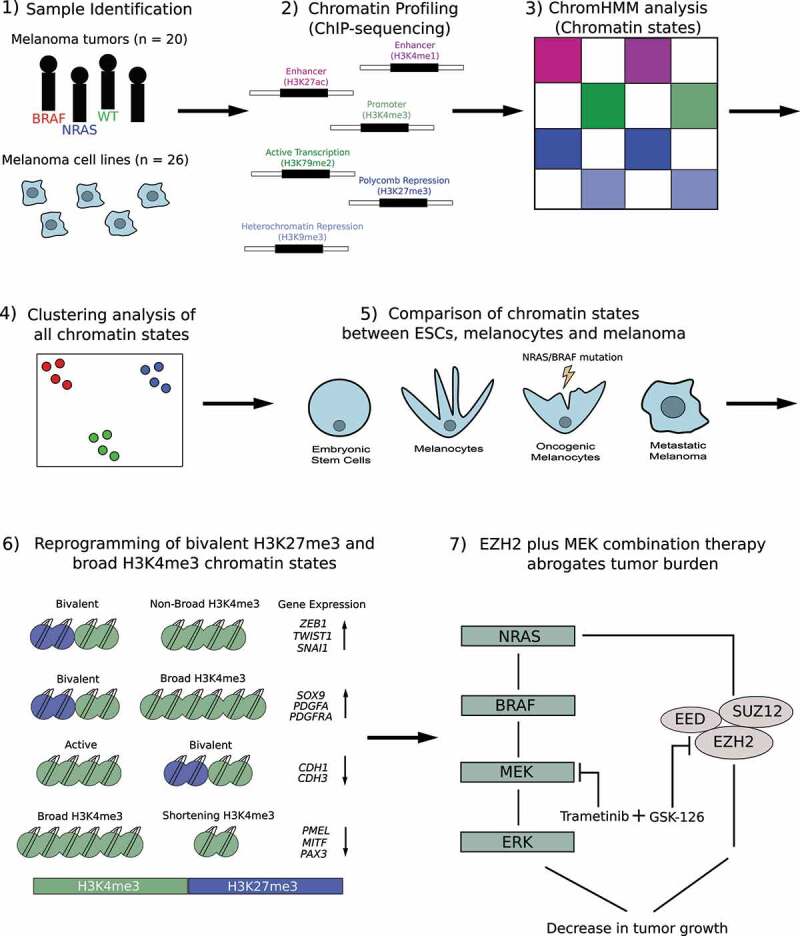
Overview of chromatin state analysis in melanoma tumor samples and cell lines. 1) Samples used in this study included 20 melanoma tumors (BRAF, NRAS, Wild-Type [WT]), 10 melanoma short-term cultures and 16 commercially available melanoma cell lines. 2). Chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-sequencing) was performed in melanoma tumors and cell lines using histone modifications for H3K4me1, H3K27ac, H3K4me3, H3K79me2, H3K27me3 and H3K9me3. 3) ChromHMM analysis was used to identify combinatorial chromatin state definitions and histone mark probabilities in 20 metastatic melanoma tumor samples. 4) Multidimensional Scaling (MDS) and differential analysis was performed for each chromatin state annotated by mutational subtype. 5) Chromatin states were compared between embryonic stem cells (ESCs), melanocytes, oncogenic melanocytes harboring an NRAS- or BRAF-mutation and melanoma samples harboring an NRAS- or BRAF-mutation. 6) Model describing potential mechanism in which switches of bivalent H3K27me3 domains on Epithelial to Mesenchymal Transition Transcription Factor (EMT-TF) and invasive genes, and shortening of H3K4me3 domains on melanocyte regulatory and proliferative genes, controls their expression during the transition to an invasive state. 7) Model describing potential therapeutic strategy for targeting the Mitogen-activated protein kinase (MEK) pathway (trametinib) and Enhancer of Zeste Homolog 2 (EZH2) (GSK-126) to block tumor burden in NRAS-mutant melanoma
