Figure 3.
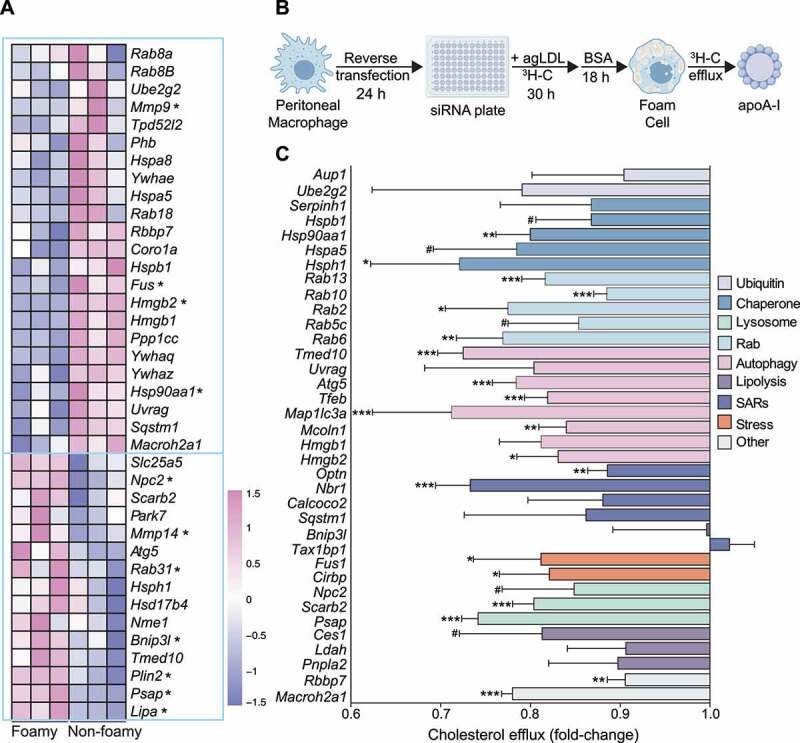
Lipophagy candidate genes are dysregulated in atherosclerosis and regulate foam cell cholesterol efflux. (A) Bulk RNA-seq of atherosclerotic foam cells from apoe−/− mice fed a Western diet for 28 weeks. Genes denoted by an asterisk are significantly different between macrophage populations (P < 0.05) and heatmap color values correspond to the relative expression (Z-score-transformed RPKM values) of each gene across the six samples. Data was acquired from Kim et al. [30] (B) Schematic for reverse transfection of mouse peritoneal macrophages. (C) Efflux of 3H-cholesterol (x-axis) from agLDL-loaded mouse peritoneal macrophages transfected with siRNAs against indicated target mouse genes (y-axis) grouped under shared functional annotations: ubiquitination machinery (Ubiquitin), molecular chaperones (Chaperone), lysosome function (Lysosome), Rab proteins (Rab), Autophagy regulators (Autophagy), regulators of neutral lipolysis (Lipolysis), selective autophagy receptors (SARs), response to cell stress (Stress) and other. Data are the mean ± s.e.m. of four independent experiments. #P < 0.1, *P< 0.05, **P< 0.005, ***P< 0.0005
